
MIXTURES OF GAUSSIAN DISTRIBUTIONS UNDER LINEAR
DIMENSIONALITY REDUCTION
Ahmed Fawzi Otoom, Oscar Perez Concha and Massimo Piccardi
Faculty of Engineering and IT, University of Technology, Sydney (UTS), Sydney, Australia
Keywords:
Dimensionality reduction, Linear transformation, Random projections, Mixture models, Object classification.
Abstract:
High dimensional spaces pose a serious challenge to the learning process. It is a combination of limited
number of samples and high dimensions that positions many problems under the “curse of dimensionality”,
which restricts severely the practical application of density estimation. Many techniques have been proposed
in the past to discover embedded, locally-linear manifolds of lower dimensionality, including the mixture of
Principal Component Analyzers, the mixture of Probabilistic Principal Component Analyzers and the mixture
of Factor Analyzers. In this paper, we present a mixture model for reducing dimensionality based on a linear
transformation which is not restricted to be orthogonal. Two methods are proposed for the learning of all
the transformations and mixture parameters: the first method is based on an iterative maximum-likelihood
approach and the second is based on random transformations and fixed (non iterative) probability functions.
For experimental validation, we have used the proposed model for maximum-likelihood classification of five
“hard” data sets including data sets from the UCI repository and the authors’ own. Moreover, we compared the
classification performance of the proposed method with that of other popular classifiers including the mixture
of Probabilistic Principal Component Analyzers and the Gaussian mixture model. In all cases but one, the
accuracy achieved by the proposed method proved the highest, with increases with respect to the runner-up
ranging from 0.2% to 5.2%.
1 INTRODUCTION
In the pattern recognition literature, it is widely rec-
ognized that high dimensional spaces cause particular
difficulties in designing a classifier. One of the rea-
sons is that, in many applications, data points are rep-
resented in a high dimensional space when intrinsi-
cally they lie in a manifold of much lower dimension-
ality. Secondly, high dimensions imply high number
of parameters which makes the computational task of
manipulating and inverting large matrices too expen-
sive. Thirdly, the behaviour and interpretability of
concepts acquired for low dimensionality spaces are
not always generalized to spaces of many dimensions.
Finally, the exponential growth for the need of train-
ing samples with the increasing number of dimen-
sions. The combination of all these severe difficul-
ties that can be present in high dimensional spaces is
called the “curse of dimensionality” (Bellman, 1961).
In order to mollify the curse, classification is often
preceded by a dimensionality reduction step where
the original features are combined into a significantly
smaller set.
One of the simplest and widely applied techniques
for this purpose is Principle Component Analysis
(PCA). In PCA, the data in the high dimensional
space (P − dimensional) are transformed by an
orthogonal projection into a lower dimensional
space (D − dimensional). This transformation is
performed in a way that maximizes the variance
of the projected data by computing the eigenvalue
decomposition of the data covariance matrix. In
the reduced space, clustering analysis, density mod-
eling and classification are carried out often with
higher classification accuracy than in the original
space. PCA has proved to be successful in many
applications; however, if the dimensionality of the
feature space is very high, the calculation of the
eigenvectors becomes computationally hard with
a complexity of O(nP
2
) + O(P
3
), where n is the
number of data vectors (often, the O(nP
2
) is the
dominant term). Moreover, variance maximization
may not necessarily lead to high class discrimination.
A simple yet interesting alternative approach is to use
random transformations (Kaski, 1998), (Fodor, 2002).
511
Otoom A., Perez Concha O. and Piccardi M. (2010).
MIXTURES OF GAUSSIAN DISTRIBUTIONS UNDER LINEAR DIMENSIONALITY REDUCTION.
In Proceedings of the International Conference on Computer Vision Theory and Applications, pages 511-518
DOI: 10.5220/0002844005110518
Copyright
c
SciTePress

In this approach, the original data Y is transformed
into the lower dimensional X via: X = WY , where W
is a D x P matrix (D << P) and its columns are re-
alizations of independent and identically distributed
(i.i.d) zero-mean normal variables, scaled to have unit
length (Fodor, 2002). Therefore, the complexity of
computing the random matrix is O(PD). It has been
shown that projecting the data to a random lower-
dimensional subspace yields results comparable to
conventional methods such as PCA, as long as the re-
duced dimension is sufficiently large (Kaski, 1998),
(Bingham and Mannila, 2001).
On the other hand, PCA has also been refined as
a maximum likelihood solution for a probabilistic la-
tent variable model, commonly known as Probabilis-
tic PCA (PPCA) (Tipping and Bishop, 1999b). In
PPCA, a P-dimensional observed data vector y can be
described in terms of a D-dimensional latent vector x
as:
y = W x + µ + ε (1)
where W is a P x D matrix describing a linear transfor-
mation and ε is an independent Gaussian noise with
a spherical covariance matrix σ
2
I. In this direction,
Factor analysis (FA) (Bartholomew, 1987) is closely
related to PPCA, except that the noise is assumed to
have a diagonal covariance matrix.
In recent years, there have been growing inter-
est in developing different techniques for discovering
embedded, locally-linear manifolds of lower dimen-
sionality that extend the above methods including:
mixture of PCA (Hinton et al., 1997), MPPCA (Tip-
ping and Bishop, 1999a), and mixture of FA (Ghahra-
mani and Hinton, 1997), amongst others.
In this paper, we present a novel probabilistic mix-
ture model for dimensionality reduction. Each com-
ponent of the mixture consists of a linear transforma-
tion projecting the original data onto a subspace and
a Gaussian distribution is fitted on the projected data.
This approach is inspired by a sensor fusion analogy,
where each component of the mixture is seen as a
sensor that can capture a good representation of the
original data by finding the best transformation ma-
trix to represent the data into a new reduced space,
and then, fitting a Gaussian distribution over the trans-
formed data. For this reason and for immediacy, we
have named the proposed method MLiT - mixture of
Gaussians under Linear Transformations.
One of the main novelties of our technique is that
the transformation matrices are not restricted to be or-
thogonal, and this paper explores how this will have
an effect on the final classification performance. Two
different ways are proposed to learn the model’s pa-
rameters:
i. The first approach initializes the transformation
matrices to orthogonal base vectors, and then
learns the parameters of all the transformation ma-
trices and Gaussian distributions in a maximum-
likelihood framework (which might cause the vec-
tors to adopt a non orthogonal arrangement) by
using an Expectation-Maximization (EM) algo-
rithm.
ii. The second approach, faster and less computa-
tionally expensive than the first one, assigns the
transformation matrices to random matrices and
fixes the Gaussians based on the sample mean and
covariance. According to (Kaski, 1998), in a high
dimensional space, there are a much larger num-
ber of sufficiently close to orthogonal than orthog-
onal vectors that might likely be found by carrying
out a random mapping.
The proposed technique, MLiT, is used to
learn class-conditional likelihoods for maximum-
likelihood classification of five “hard” data sets from
the UCI repository and the authors’ own. Moreover,
our model is compared against the well known MP-
PCA and the conventional Gaussian Mixture Model
(GMM).
This paper is organized as follows: Section 2
presents the proposed method (MLiT), the maximum
likelihood solution, the initialization procedure for
this solution, and the learning of the transformation’s
parameters using random matrices. Section 3 de-
scribes the experiments conducted to evaluate MLiT
over multiple data sets, comparing the results with
state-of-the-art classifiers. Finally, in section 4, we
draw our conclusions and discuss future work.
2 APPROACH AND
METHODOLOGY
In this section, we describe MLiT, a method for gen-
erating a mixture distribution in a dimensionally re-
duced space that can be useful for density modelling
and classification. We first describe the model in the
next subsection. We then present in Subsection 2.2
the maximum-likelihood solution devised to learn the
model from a set of samples. We also discuss the
initialization procedure for this solution. Finally, in
Subsection 2.3, we present another way for learning
the model’s parameters based on random matrices.
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
512

2.1 Mixture of Gaussians under Linear
Transformations (MLiT)
Let us consider a multivariate random variable, y, in
a high, P-dimensional space. We define the lower,
D-dimensional space through a compressive linear
model
x = Ωy (2)
where Ω is a D ×P real matrix, with D <= P and
typically D << P. We also posit a density function,
p(x), in x-space and consider
f (y) = p(Ωy) = p(x); (3)
f (y) is not a proper density in y-space: rather, a
probability function that repeats the probability den-
sity p(x) for all y points satisfying x = Ωy. As such,
f (y) expresses the probability of the combination of
two distributions: a distribution modelled by p(x) in
the D-dimensional subspace spanned by the rows of
Ω (the retained dimensions); and a uniform distribu-
tion along the (P −D)-dimensional subspace satisfy-
ing equation x = Ωy for any given x (the discarded di-
mensions). For instance, if p(x) is Gaussian, f (y) has
the shape of a Gaussian “ridge” i.e. a D-dimensional
Gaussian function which repeats itself along the di-
rection of x = Ωy in y-space. When referring to its
distributional properties hereafter, we will refer to this
distribution as Gaussian-uniform. Following the sen-
sor analogy, x can be seen as a view of y made avail-
able by a sensor. If the representation power of x is
adequate, it will permit to successfully study prop-
erties of y e.g. classify measurements in classes of
interest.
In general, exploiting an array of M sensors can
offer a richer representation of y than a single sensor.
By calling f (y|l) the probability function for the l-th
sensor in the array, l = 1..M, it holds that:
f (y|l) = p(Ω
l
y|l) (4)
where we have assumed that each sensor has its
own independent view of y, expressed by Ω
l
(Kittler,
1998).
Let us now assume that we have a way to estimate
a discrete distribution, p(l), stating the quality of the
l-th sensor at explaining the y sample. From Bayes
theorem, we obtain:
f (y,l) = f (y|l)p(l) = p(Ω
l
y|l)p(l) (5)
By marginalizing over l, we obtain the probability
function f (y) for the sensor array case:
f (y) =
M
∑
l=1
f (y,l) =
M
∑
l=1
f (y|l)p(l) =
M
∑
l=1
p(Ω
l
y|l)p(l)
(6)
which closely recalls the general density of a mix-
ture distribution. However, probabilities are com-
puted in subspaces spanned by linear transformations
and such transformations differ for each component.
For simplicity of treatment, we further assume that
the individual sensor densities are Gaussian, and note
α
l
= p(l), obtaining:
f (y) =
M
∑
l=1
α
l
N (Ω
l
y|µ
l
,Σ
l
) (7)
where the N (Ω
l
y|µ
l
,Σ
l
) terms are the densities in
the subspaces; means µ
l
, and covariance matrices Σ
l
are the parameters of each Gaussian component in the
l-th subspace for l = 1..M; weights α
l
are the mixing
coefficients.
Once an f (y) density is learnt for each c class,
c = 1..C, maximum likelihood classification can be
simply attained as:
c
∗
= argmax
c
( f (y|c)) (8)
We note that this model makes no attempt at posi-
tioning the subspaces over clusters of data in y-space
or minimizing reconstruction errors. As such, the
number of views is not in correspondence with the
number of clusters in the sample set. Rather, each
view is justified by a good likelihood fit i.e. providing
high within-class invariance.
2.2 Maximum Likelihood (ML)
Solution
We propose maximum likelihood as one way for
jointly finding the parameters θ
l
= {α
l
,µ
l
,Σ
l
,Ω
l
}
M
l=1
.
To this aim, we consider a set of i.i.d. observations,
Y =
{
y
i
}
i=1..N
, in the high dimensional space. Our
goal is then that of finding values for parameters of
(7) maximizing likelihood
L(θ) = p(Y |θ) =
N
∏
i=1
f (y
i
) =
N
∏
i=1
(
M
∑
l=1
α
l
N (Ω
l
y
i
|µ
l
,Σ
l
))
(9)
where θ = {Ω
l
,α
l
,µ
l
,Σ
l
} and l = 1..M. As usual
in similar cases, rather than attempting maximization
of (9) directly, we adopt an EM approach. This re-
quires positing the existence of a set of discrete, M-
valued latent variables, Z =
{
z
i
}
i=1..N
, whose mini-
mum requirement is that the expression of joint prob-
ability function f (y
i
,Z) be simpler than f (y
i
) itself.
MIXTURES OF GAUSSIAN DISTRIBUTIONS UNDER LINEAR DIMENSIONALITY REDUCTION
513

The target for maximization is the expected value of
the complete-data log-likelihood,
Q(θ,θ
g
) =
∑
Y
[ln(p(Y,Z|θ)p(Z|Y,θ
g
)] (10)
where θ and θ
g
represent the new and old parame-
ters in the EM iterations, respectively. In (10), Z rep-
resents a single realization of the entire set of the la-
tent variables and the summation extends over all its
possible M
N
values. The whole derivation has been
presented by the authors in a previous work.
The E-step computes p(z
i
= l|y
i
,θ
g
), or p(l|y
i
,θ
g
)
for brevity, which is the responsibility of the l-th com-
ponent for the y
i
sample (Bishop, 2006).
p(l|y
i
,θ
g
) =
α
g
l
N (Ω
l
y
i
|µ
g
l
,Σ
g
l
)
∑
M
k=1
α
g
k
N (Ω
k
y
i
|µ
g
k
,Σ
g
k
)
(11)
Maximizing (10) leads to the following M-step for
the parameters:
α
l
=
1
N
N
∑
i=1
p(l|y
i
,θ
g
) (12)
µ
l
=
∑
N
i=1
Ω
l
y
i
p(l|y
i
,θ
g
)
∑
N
i=1
p(l|y
i
,θ
g
)
(13)
Σ
l
=
Σ
N
i=1
(Ω
l
y
i
−µ
l
)(Ω
l
y
i
−µ
l
)
T
p(l|y
i
,θ
g
)
∑
N
i=1
p(l|y
i
,θ
g
)
(14)
For the maximization of Ω
l
, we considered this
matrix as P,D ×1 column vectors, Ω
l
= (w
j
)
l
, j =
1..P, and we update it column by column, rather than
the whole matrix at once. Therefore, the re-estimation
formula for (w
1
)
l
is the following:
(w
1
)
l
=
=
∑
N
i=1
(−(w
g
2
)
l
y
i2
−... −(w
g
P
)
l
y
iP
+ µ
g
l
)y
i1
p(l|y
i
,θ
g
)
∑
N
i=1
y
2
i1
p(l|y
i
,θ
g
)
(15)
where N is the number of samples, (w
g
j
)
l
, j = 2..P
are the other columns’ “old” values, θ
g
represent the
model’s old parameters.
Two important issues may occur as a result of the
projection step:
i. the component densities across the mixture model
and across different classes, can be different in
scale. By this, we mean that the linearly trans-
formed space (the x-space) does not have a de-
fined scale; therefore, likelihood p(x) can be made
arbitrarily larger or smaller by changes to the scale
of x.
ii. as a consequence of this and the maximum-
likelihood target, the scale of x may tend to 0
along iterations to endorse high values of p(x). In
turn, this implies that the projection matrix may
also tend to zero (an undesirable solution that we
call degenerate or singular hereafter).
In order to avoid these problems, we propose the
normalization of the projection matrix at each step of
the EM algorithm; henceforth, we refer to this method
as MLiT (Normalized) or MLiT (N). By equating the
concept of norm to that of scale, this will make the
densities of equal scale across the different compo-
nents, and also across different classes. Further, this
will avoid Ω
l
reaching the degenerate solution and act
as a likelihood regularization. Therefore, after each
EM step, we normalize Ω
l
as follows:
Ω
l
=
Ω
l
Norm(Ω
l
)
(16)
We have tried several norms (L1, L2, Infinity and
Frobenius), with the Frobenius norm providing the
highest and most stable results. Therefore, in the fol-
lowing, we report results based on this norm:
Frobenius norm(Ω
l
) =
q
∑
diag(Ω
T
l
Ω
l
) (17)
Thus, MLiT (N) searches for possible solutions
over the likelihood space. Every time a solution is
provided by the maximization step of EM, we nor-
malize the projection matrix Ω
l
in order to keep it
on an equal scale. The expectation-maximization
steps become therefore expectation-maximization-
normalization steps. An obvious disadvantage of this
approach is that the new normalized solution might
or might not have higher likelihood than the previ-
ous normalized solution. For this reason, we monitor
the evolution of the likelihood along the iterations and
elicit ad-hoc convergence criteria.
2.2.1 Initialization Phase
In EM, the parameter values traversed along the it-
erations and the likelihood value achieved at conver-
gence may strongly depend on the parameters’ initial
values. For our approach, we choose to apply a deter-
ministic initialization to ensure repeatable results at
each run. Namely, we decided to initialize the pro-
jection matrix, Ω, by the orthonormal transformation
provided by PCA, selecting either the largest or the
smallest eigenvectors (i.e. the eigenvectors associ-
ated with the largest and smallest eigenvalues, respec-
tively).
Projecting the data by the largest eigenvectors
transforms them into a space where their variance
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
514

is maximized and, under the hypothesis that their
distribution be Gaussian, the likelihood is minimum
amongst all orthonormal projections (Bolton and
Krzanowski, 1999), and therefore forcing the EM to
explore a large region of the parameter space before
convergence. Conversely, projecting them with the
smallest eigenvectors transforms them into a space
where their variance is minimized and likelihood is
maximum.
In our experiments, we noticed that there is no cer-
tain initialization method that always provide the best
classification accuracy. Thus, we experiment with
both methods and choose the one providing better ac-
curacy results.
As the data per class are projected to each of the
components, the remaining initial parameters of the
EM algorithm are chosen as follows:
• The initial mean µ
l
and covariance matrix Σ
l
of
each component will be the sample mean and
covariance computed directly from the projected
data of each component, x
l
:
– µ
l
=
1
N
∑
N
n=1
x
ln
– Σ
l
=
1
N−1
∑
N
n=1
(x
ln
−µ
l
)(x
ln
−µ
l
)
T
• The initial priors α
l
for l = 1..M, are chosen to be
equal across all the components:
– α
l
=
1
M
2.3 Random Transformations Solution
In this subsection, we present another method for
learning the transformation matrix, Ω
l
, based on us-
ing random matrices (MLiT (R)). The main idea is to
use random matrices of Gaussian distributed elements
and with unit length columns. This idea is moti-
vated by the Johnson-Lindenstrauss lemma (Johnson
and Lindenstrauss, 1984): if points in a feature space
Y (P-dimensional) are projected onto a randomly se-
lected subspace of suitable high dimension D, then the
distances between the points are approximately pre-
served if D is large enough.
<(||φ(y
i
) −φ(y
j
)||
2
D
−||y
i
−y
j
||
2
P
)
2
>
φ
≤
≤
2
D
||(y
i
) −(y
j
)||
4
P
(18)
where || ||
P
and || ||
D
denote the Euclidean dis-
tance norms in V
P
and V
D
, respectively, and <>
φ
is
the average over all possible isotropic random choices
for the unit vectors defining the random mapping φ.
In our case, we chose the elements of the Ω
l
to
be drawn from a zero-mean normal distribution with
a variance of 1/
√
P, where P is the original space
dimension. The columns of Ω
l
are then normalized
to be unitary vectors. Moreover, we decided to have
the same transformation matrices across all classes.
Thus, with the above settings, the scale of the trans-
formation matrices is comparable.
After transforming the data of each class as: X
l
=
Ω
l
Y , we fix a Gaussian distribution over the trans-
formed data and the mixture parameters are as fol-
lows:
• The priors α
l
for l = 1..M, are chosen to be equal
across all the components.
• The final mean µ
l
and covariance matrix Σ
l
of
each component are the sample mean and covari-
ance, computed directly from the projected data
of each component, x
l
.
3 EXPERIMENTS AND ANALYSIS
The empirical evaluation of a classifier’s accuracy re-
quires extensive testing over multiple data sets and
a comparative analysis with existing, state-of-the-art
classifiers. To this aim, in this section we present de-
tails on the data sets used and experiments conducted.
3.1 Data Sets
We evaluate the proposed method on five data sets,
four of which are selected from the UCI Machine
Learning Repository (Asuncion and Newman, 2007),
and are widely used by the pattern recognition com-
munity for evaluating learning algorithms. These four
data sets are the Vehicle data set, Wisconsin Diagnos-
tic Breast Cancer data set (WDBC), Wisconsin Prog-
nostic Breast Cancer (WPBC) data set, and Optical
Handwritten Digits data set (OpticDigit). The last
data set, named Public Premises Video Surveillance
data set (PPVS), was collected by the authors them-
selves.
The Vehicle data set involves classification of a
given silhouette as one of four types of vehicles,
namely, “bus”, “Opel”, “Saab” and “van”. The ve-
hicle silhouettes are described by various shape mea-
surements. The rationale for choosing this data set is
that it is the most similar in the UCI repository to our
own data set and can offer a comparative insight into
the method’s performance. The WDBC and WPBC
data sets contain various shape features from images
of fine needle aspirates (FNA) of breast mass for diag-
nosis and prognosis of breast cancer. The OpticDigit
data set is based on rescaled bitmaps of handwritten
digits: the original 32x32 black and white bitmaps
are divided into non-overlapping blocks of 4x4 pixels
and the number of ‘on’ pixels counted in each block,
MIXTURES OF GAUSSIAN DISTRIBUTIONS UNDER LINEAR DIMENSIONALITY REDUCTION
515

Table 1: Comparative summary of the data sets used.
Data set # Features # Instances # Classes
Vehicle 18 846 4
OpticDigit 64 5620 10
WDBC 30 569 2
WPBC 33 198 2
PPVS 44 600 4
resulting in a 64-dimensional feature vector of homo-
geneous features. The Public Premises Video Surveil-
lance data set (PPVS) is based on video footage pro-
vided by an industrial partner. It involves classifica-
tion of an object in a video surveillance environment
into one of four classes: “trolleys”, “bags”, “single
persons”, and “groups of people”. The images of
these objects have been clipped from video footage
acquired at a number of airports and train stations
world-wide. The feature set consists of statistics of
various local features such as line segments, circles,
corners, and global shape descriptors such as fitted
ellipses and bounding boxes. This feature set is de-
scribed in detail in (Otoom, 2007).
As we can conclude from the previous paragraphs
and the data displayed in Table 1, there are major
differences between these five data sets in terms of
the nature of data and application context, number
of instances available, number of features extracted,
types of features used for representation and number
of classes. Therefore, the chosen data sets offer a suit-
able basis for comparative analysis.
3.2 Experiments
In this subsection, we present classification results for
the proposed method on the five aforedescribed data
sets. We compare the performance of our approach
with that of mixture of PPCA (MPPCA) and Gaussian
mixture model (GMM). Experiments with these clas-
sifiers were carried out in MATLAB by setting all tun-
able parameters in the most genuine way to achieve
the highest performance. We summarize below the
main parameters, and in Table 2, we report the values
that achieved the best accuracy results. The parame-
ters are as follows:
• GMM: There is one main parameter, the number
of the GMM components (M).
• MPPCA: There are two main parameters, the
number of reduced dimensions (D), and the num-
ber of mixture components (M).
For MLiT, the parameters to adjust are selected as
follows:
• The number of the mixture components (M) and
reduced dimensions (D) were manually selected
as reported in Table 2.
• For MLiT (N):
– The initial transformation matrices for each
class, Ω
[0]
, were computed by using either the
smallest or the largest consecutive eigenvectors
of the covariance matrix of the original data.
For example, in the case of largest eigenvec-
tors, two components per class (M = 2), and a
reduced space of three dimensions (D = 3), we
select the three first eigenvectors for Ω
[0]
1
and
the eigenvectors between the third and the fifth
for Ω
[0]
2
.
– Initial transformed data: X
[0]
l
= Ω
[0]
l
Y , l = 1..M.
– Initial means, µ
[0]
l
, and variances, Σ
[0]
l
, l = 1..M:
from the initial transformed data.
– Equal initial priors for all components, α
[0]
l
=
1
M
, l = 1..M.
• For MLiT (R):
– Ω
l
is chosen randomly from a 1-D zero-mean
Gaussian distribution with a variance of 1/
√
P.
– The priors α
l
for l = 1..M, are chosen to be
equal across all the components: α
l
=
1
M
.
– The mean µ
l
and covariance matrix Σ
l
of each
component are the sample mean and covari-
ance, computed directly from the projected data
of that component, x
l
.
As stopping criteria, for MLiT (N) the normal-
ization step does not guarantee a monotonic increase
in the likelihood; hence, we elicit an ad-hoc criteria
for stopping by running the EM algorithm for 50 it-
erations and choosing that delivering the maximum
accuracy by cross-validation. For MPPCA, we ob-
served that the accuracy stabilized after 200 iterations.
For GMM, instead, accuracy stabilization was empir-
ically achieved after 50 iterations. For validation, we
have chosen 5-fold cross-validation since it offers a
good trade off between the large bias of the hold-out
method and the large variance of the leave-one-out
method (Breiman and Spector, 1992). This implies
randomly partitioning the data set into five disjoint
subsets, training the classifier with four and using the
last for testing. Classification accuracy is averaged
over five runs by using, in turn, each fold for testing.
We express classification accuracy simply as the per-
centage of correctly classified instances with respect
to their total number:
accuracy =
number of correctly classified samples
total number of samples
(19)
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
516
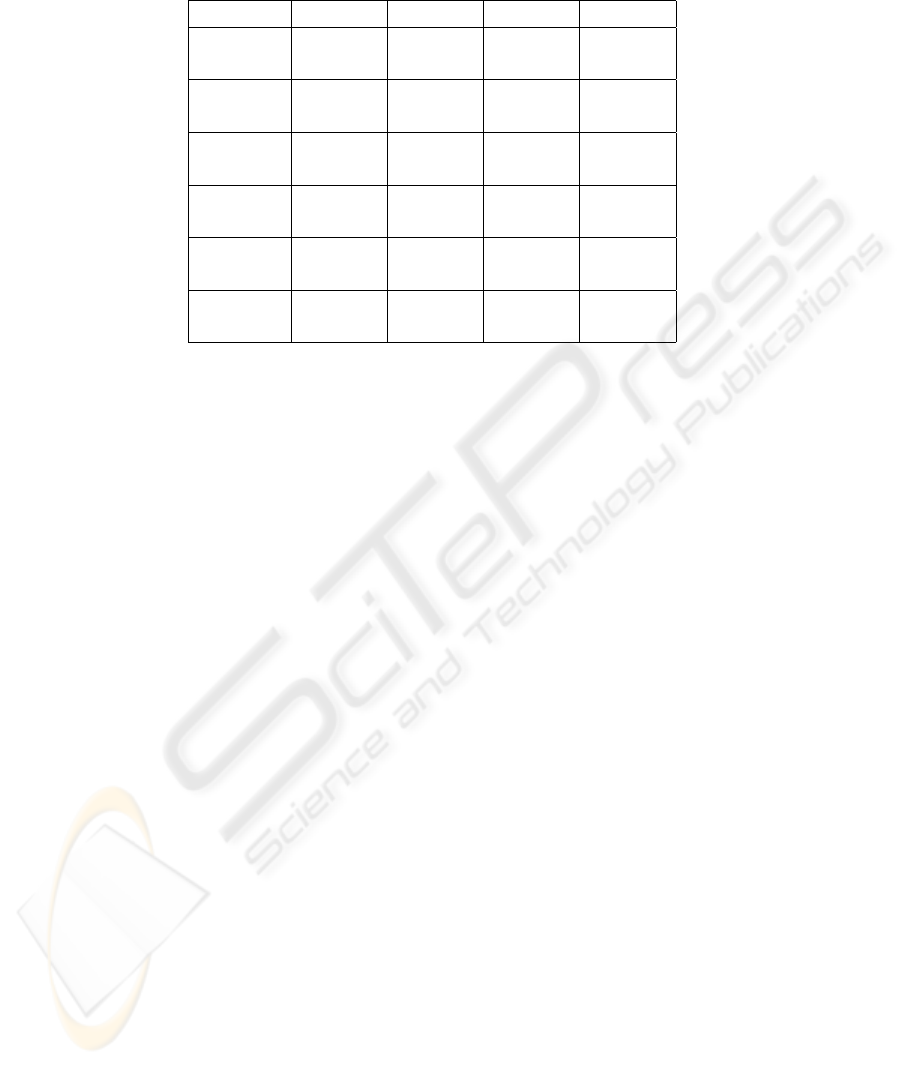
Table 2: Results for 5-fold CV in terms of accuracy (%) and standard deviation, on five data sets and across different classifiers,
with the highest indicated in boldface font. For each dataset, the first row presents the main parameters’ values for different
classifiers, and the second row shows the achieved accuracy (%).
Classifier MLiT (N) MLiT (R) MPPCA GMM
Parameters D M D M D M M
Dataset
PPVS 33 1 30 2 25 2 2
(%) 76.7 ±2.4 78.5 ±2.0 73.5 ±4.0 73.3 ±2.1
Vehicle 14 2 18 2 10 2 2
(%) 85.6 ±1.9 84.3 ±2.1 83.6 ±1.3 82.8 ±1.9
OpticDigit 29 2 35 5 16 1 2
(%) 98.4 ±0.3 98.3 ±0.3 98.6 ±0.4 96.9 ±0.3
WDBC 18 1 20 4 20 2 2
(%) 96.1 ±1.7 95.9 ±1.9 94.7 ±1.7 95.9 ±1.8
WPBC 4 4 25 2 15 4 4
(%) 77.4 ±1.1 76.9 ±1.8 76.9 ±0.0 75.9 ±1.4
It is important to note that, in the following, we
report the classification results in terms of two sta-
tistical measures: average accuracy over the various
runs, and standard deviation. However, we chose the
average accuracy as the main measure for comparing
the different classifiers; nevertheless, the standard de-
viation is an important measure for the precision of
the classification accuracy, and it can be considered
together with the accuracy for a better estimate of the
classification performance.
Table 2 reports the best results of 5-fold cross-
validation on the various data sets and across the com-
pared classifiers. We note that in this table, for MLiT
(N), all results are obtained with largest eigenvectors
initialization except the cases of Vehicle and WPBC
data sets. It is clear from this table that, in all cases
(except the PPVS data set), MLiT (N) has slightly
outperformed the performance of MLiT (R), proving
that the maximum likelihood solution can be a bet-
ter learning method in comparison to that of learn-
ing based on random matrices. However, the mar-
gin of improvements is not very high, which indi-
cates that the random transformations solution can de-
liver promising results with less computation. We can
also note from Table 2 that the performance of MLiT
outperformed that of MPPCA on four out of the five
compared data sets with improvements ranging from
0.5% to 5.0%, proving the strength of MLiT against
a state-of-the-art classifier (MPPCA has slightly out-
performed MLiT by 0.2% only on the OpticDigit data
set). Moreover, we can note that, in all cases, the per-
formance of MLiT outperformed that of GMM with
improvements ranging from 0.2% to 5.2%. This il-
lustrates the ability of MLiT in overcoming the curse
of dimensionality and providing better performance
in the reduced space. MPPCA has also provided bet-
ter classification results than GMM on majority of the
data sets.
Overall, the experiments on the five data sets pre-
sented in this section showed that MLiT reported
higher experimental accuracy over both compared
classifiers (except the case of OpticDigit where MP-
PCA slightly outperformed MLiT). Interpretation of
accuracy results in high dimensional spaces is not im-
mediate. In the case of the ML solution, we lean
to attribute these improvements in accuracy to the
Gaussian-uniform distribution property of focussing
on invariant features. This permits the building
of compact models that have proved discriminative
when used with the Bayes inversion rule, while it
introduces elements of robustness since outliers are
ousted to the discarded dimensions during training
as much as possible. The non-orthogonality of the
transformation adds further degrees of freedom to the
model. However, this feature seems to be used only to
a limited extent since the maximum accuracy is often
achieved during the very first iterations of EM, when
the transformation only mildly deviates from orthog-
onality. In these terms, both the maximum likelihood
and random solutions are often close to orthogonal
transformations.
4 CONCLUSIONS
In this paper, we have presented a method for lin-
ear dimensionality reduction within mixture distribu-
tions. The model that we have proposed for the class-
conditional likelihood is a mixture of Gaussian distri-
butions under linear transformations (7). This model
MIXTURES OF GAUSSIAN DISTRIBUTIONS UNDER LINEAR DIMENSIONALITY REDUCTION
517

equates to a uniform distribution along the discarded
dimensions and a full Gaussian model along the re-
tained dimensions.
It is important to contrast this model properly to
the several existing methods for linear dimensionality
reduction in mixture models such as mixtures of PCA,
PPCA, and FA. One of the main points of difference is
that the linear transformation is not restricted to be or-
thogonal. Further, the linear model adopted, x = Ωy,
does not assume additive noise models and makes x
observable. On the ground of that, we can evaluate
density N (Ωy|µ,Σ) = N (x|µ,Σ) directly in x-space.
For learning the model, we have presented two differ-
ent methods; the first method learns the model’s pa-
rameters in a maximum likelihood framework (MLiT
(N)). Normalization is proposed as a way to regular-
ize this solution. Thus, a common scale is imposed
to all the transformations and a singularity problem
is avoided. Another simple yet powerful method for
learning the model’s parameters can be based on ran-
dom matrices (MLiT (R)). This method has offered
promising and computationally feasible results. How-
ever, the maximum likelihood solution delivered bet-
ter accuracy results in majority of the data sets sug-
gesting that it can be a better way for learning the
model’s parameters.
The experimental performance of MLiT has
proved to outperform that of MPPCA and GMM in al-
most all cases with improvements ranging from 0.2%
to 5.2% compared to the runner-up. The only case
where MLiT did not deliver the best accuracy is on
the OpticDigit data set where it was slightly outper-
formed by MPPCA by 0.2%. In addition to visual ob-
ject classification, the proposed method permits gen-
eral application for density modeling and classifica-
tion of other continuous numerical data requiring di-
mensionality reduction. Moreover, its re-estimation
formulas can be easily extended to suit boosting and
other weighted maximum likelihood targets and adapt
to a variety of pattern recognition frameworks.
ACKNOWLEDGEMENTS
The authors wish to thank the Australian Research
Council and iOmniscient Pty Ltd that have partially
supported this work under the Linkage Project fund-
ing scheme - grant LP0668325.
REFERENCES
Asuncion, A. and Newman, D. (2007). UCI machine learn-
ing repository.
Bartholomew, D. J. (1987). Latent Variable Models and
Factor Analysis. Charles Griffin & Co. Ltd., London.
Bellman, R. (1961). Adaptive control processes - A guided
tour. Princeton University Press, Princeton, New Jer-
sey.
Bingham, E. and Mannila, H. (2001). Random projection in
dimensionality reduction: applications to image and
text data. In Proceedings of the 7th ACM SIGKDD In-
ternational Conference on Knowledge Discovery and
Data Mining (KDD-2001), pages 245–250.
Bishop, C. M. (2006). Pattern Recognition and Machine
Learning. Springer.
Bolton, R. J. and Krzanowski, W. J. (1999). A characteri-
zation of principal components for projection pursuit.
The American Statistician, 53(2):108–109.
Breiman, L. and Spector, P. (1992). Submodel selection and
evaluation in regression: The x-random case. Interna-
tional Statistical Review, 60(3):291–319.
Fodor, I. (2002). A survey of dimension reduction
techniques. Technical Report UCRL-ID-148494,
Lawrence Livermore National Laboratory.
Ghahramani, Z. and Hinton, G. (1997). The EM algorithm
for mixtures of factor analyzers. Technical Report
CRG-TR-96-1, University of Toronto.
Hinton, G. E., Dayan, P., and Revow, M. (1997). Modeling
the manifolds of images of handwritten digits. IEEE
Transactions on Neural Networks, 8(1):65–74.
Johnson, W. B. and Lindenstrauss, J. (1984). Extensions
of lipschitz mappings into a hilbert space. In Confer-
ence in modern analysis and probability, Contempo-
rary Math, volume 26, pages 189–206.
Kaski, S. (1998). Dimensionality reduction by random
mapping: Fast similarity computation for clustering.
In Proceedings of IJCNN’98, International Joint Con-
ference on Neural Networks, volume 1, pages 413–
418. IEEE Service Center.
Kittler, J. (1998). Combining classifiers: A theoreti-
cal framework. Pattern Analysis and Applications,
1(1):18–27.
Otoom, A. e. a. (2007). Towards automatic abandoned ob-
ject classification in visual surveillance systems. In
Asia-Pacific Workshop on Visual Information Process-
ing, pages 143–149, Tanian, Taiwan.
Tipping, M. E. and Bishop, C. M. (1999a). Mixtures of
probabilistic principal component analyzers. Neural
Computation, 11(2):443–482.
Tipping, M. E. and Bishop, C. M. (1999b). Probabilistic
principal component analysis. Journal of the Royal
Statistical Society: Series B (Statistical Methodol-
ogy),, 61(3):611–622.
VISAPP 2010 - International Conference on Computer Vision Theory and Applications
518
