
EFFICIENT SEMI-AUTOMATIC MAINTENANCE OF MAPPING
BETWEEN ONTOLOGIES IN A BIOMEDICAL ENVIRONMENT
Imen Ketata, Riad Mokadem, Franck Morvan and Abdelkader Hameurlain
Institut de Recherche en Informatique de Toulouse IRIT, Paul Sabatier University
118 Route de Narbonne F-31062, Toulouse Cedex 9, France
Keywords: Biomedical Data Sources, Data Integration, Ontology, Mapping.
Abstract: In dynamic environments like data management in biomedical domain, adding a new element (e.g. concept)
to an ontology O
1
requires significant mapping creations between O
1
and the ontologies linked to it. To
avoid this mapping creation for each element addition, old mappings can be reused. Hence, the nearest
element w to the added one should be retrieved in order to reuse its mapping schema. In this paper, we deal
with the existing additive axiom which can be used to retrieve this w. However, in such axiom, the usage of
some parameters like the number of element occurrence appears insufficient. We introduce the calculation
of similarity and the user’s opinion note in order to have more precision and semantics in the w retrieval. An
illustrative example is presented to estimate our contribution.
1 INTRODUCTION
Computer science applications in biomedicine
accumulate important data volume by the
establishment of huge network of heterogeneous and
autonomous data sources distributed all over the
world. Every day new sources are published.
Consequently, the major challenge is to present a
view of these sources to help users accessing and
integrating them. There are three approaches in the
literature: navigational (Davidson et al., 1995), data
warehouse (Hammer and Schneider, 2003) and
mediation (Hernandez and Kambhampati, 2004).
The navigational approach is not widely used since
the difficulty of keeping up to date its entire static
links on which user leans to navigate between the
various web pages. Data warehouse deals with
copying and updating all the data in all sources and
integrating them into a local warehouse. However,
this is not easy, especially in a domain where
sources are added almost in a daily way with rather
important volumetric, e.g. the biomedical domain.
Finally, the mediation approach surmounts the
difficulties confronted in the previous approaches,
particularly those of update, by using the view and
global schema principle with adopting the mediator-
wrapper architecture.
In previous studies the principle of mediation
often relies on a global schema. Whereas, when such
type of schema is used, resolving several synonymy
and polysemy problems, which are more crucial in
the biomedical domain, is a difficult target. This
problem brings forth other methods which describe
the source contents in a more explicit way to assess
to its meanings, e.g. ontologies (Jonquet et al.,
2008). Developing a standard and global ontology is
the perfect solution for presenting (schematically) all
sources in a unique and homogeneous format.
However, until today, developing such ontology has
been seen as a very difficult task (Aumueller et al.,
2005). Hence, a solution emerged: the domain
ontology (Karmakar, 2007), so that, each domain is
presented by a domain ontology.
To move from one domain to another, we have to
go by some correspondence relations between them.
Establishing this type of communication means
creating connections (correspondences) between
domain ontologies what we call: mapping (Choi et
al., 2006, Aumueller et al., 2005 and Drumm et al.,
2007). In dynamic environments, it is very difficult
to maintain this mapping up to date.
Given the dynamic nature of biomedical
environment and the very rapid evolution of data
amount, creating and maintaining this mapping up to
date manually is expensive, slow and complicated to
achieve. Hence, researchers have studied to
automate this process. However, creating and
257
Ketata I., Mokadem R., Morvan F. and Hameurlain A. (2010).
EFFICIENT SEMI-AUTOMATIC MAINTENANCE OF MAPPING BETWEEN ONTOLOGIES IN A BIOMEDICAL ENVIRONMENT.
In Proceedings of the 12th International Conference on Enterprise Information Systems - Databases and Information Systems Integration, pages
257-262
DOI: 10.5220/0002905702570262
Copyright
c
SciTePress

maintaining the mapping with a full automatic way
without expert’s intervention is hard to realize
(Aumueller et al., 2005). Indeed, semi-automatic
solutions can automate mapping updating task with
reduced expert’s effort. Several studies have
emerged in this context, such as COMA++
(Aumueller et al., 2005) and QuickMig (Drumm et
al., 2007).
Keeping the mapping up to date, after an element
addition (e.g. concept), requires creating mapping
between the newly added element and existing ones.
Reusing existing mappings can avoid the creation of
a new mapping at each element addition. For this
reason, it seems natural to seek the most similar
element to that added in order to reuse its mapping.
Having an effective selection of this similar element
means having essentially a high semantic level in
ontologies and its mapping schemas (Drumm et al.,
2007 and Choi et al., 2006). This means in others
words, introducing concepts and mapping relations
in a more explicit and accurate way. To deal with
this semantic, some researchers rely on similarity
between elements (Couto et al., 2007). Other
approaches deal with the similarity between graphs
(Melnik et al., 2002), semantic similarity relations
between descriptions in ontologies (Hakimpour and
Geppert, 2002) and using the word similarity to
semi-automatic element generation in ontologies
(Weeds and Weir, 2005). The first two approaches
are used for schema and relation similarities. The
third one, proposed in (Weeds and Weir, 2005) is the
most appropriate to deal with similarity between two
concepts (words). It relies on theorem called
additive axiom to retrieve the nearest word to a
newly added one. This axiom calculation is based
primarily on the common features between the
added and the old word and secondly on the
importance of the old word. This importance is
measured by the number of word occurrence
(appearance). Although the occurrence number is
necessary to calculate the word importance, it is not
sufficient in some cases since there are several
words appearing many times while they do not have
an importance. Thus, addition of other parameters in
the word importance calculation should be
examined. In this paper, we propose to add two
parameters in order to have more precision and
semantics in the calculation of the word importance
in additive axiom. Precisely, we introduce the
similarity calculation for more accuracy in the
additive axiom results. We involve also the opinion
of the users as they might be experts in the
biomedical domain (e.g. doctor, biologist). So we
can benefit from the user’s experience and
knowledge to evaluate the importance of a word.
Subsequently, the addition of these two parameters
(similarity calculation and user’s opinion) can lead
to a more accurate and meaningful selection for the
nearest word to the added one for a better semi-
automatic mapping maintenance.
In the second section, we define basic concepts
in data integration for the biomedical domain. In the
third section, we introduce our contribution. After
that, to validate our work we evaluate it by an
illustrative example. The final section contains
concluding remarks and future perspectives.
2 BASIC CONCEPTS IN DATA
INTEGRATION FOR THE
BIOMEDICAL DOMAIN
The very fast evolution of the data sources in a
dynamic environment generates several complex
problems which are much more complicated in the
biomedical field since the huge volumetric of data
sources and their number evolving over the time in a
very fast way. This requires a big storage capacity
infrastructure, e.g. Grid.
The use of ontology (which is more suitable than
a global schema, as showed earlier), is a solution in
which several researchers have been interested
(Hernandez and Kambhampati, 2004). However, it is
very difficult to have a standard ontology
representing the entire biomedical domain. One
solution emerges, that is to present (conceptualize)
each sub-domain of biomedical domain by a domain
ontology (Karmakar, 2007). In this paper, we
represent each biomedical sub-domain by a domain
ontology. To integrate data sources related at each
domain ontology, mediation is the most suitable
approach. Therefore, at each domain ontology, a
number of mediators is associated. The passage from
one mediator to another in the same sub-domain is
not a problem since the communication between
them is established through common rules. This is
not the case with two mediators belonging to
different sub-domains. Besides, to be able to link
semantically concepts in this last case, we must
establish the mapping between domain ontologies.
Various research works have focused on this context
dealing with the semantic heterogeneity. In this
paper, we are especially interested in the work of
(Weeds and Weir, 2005) which helped to maintain
the ontology mapping.
The mapping involves integrating a set of
independently developed schemas and/or ontologies
ICEIS 2010 - 12th International Conference on Enterprise Information Systems
258

into a single one (Drumm et al., 2007 and Madhavan
et al., 2001). There are two kinds of mapping: (i) the
mapping between an ontology and the schema or the
local ontology related to a data source (Choi et al.,
2006 and Silva and Rocha, 2003) and (ii) the
mapping between ontologies (Weeds and Weir, 2005
and Doan et al., 2002). The first type allows
communication between an ontology and its sources.
In the second type, ontologies are connected two by
two without taking into account the type of the
topology allowing so more autonomy.
At each new element addition w’ (concept), an
appropriate mapping must be established. Reusing
old mapping related to the nearest element w to the
added one w’ can avoid recreating this mapping at
each insertion as we noted earlier. So, the proximity
between the added element and existing ones must
be calculated to find the nearest one. Diverse studies
are interested in this problem. We quote for example
those which are based on the similarity calculation:
(Melnik et al., 2002, Hakimpour and Geppert, 2002
and Weeds and Weir, 2005). This last work deals
with the distributional similitude and the semantic
similarity between elements. It is exactly interested
in the applicative domain for automatic ontology
generation (e.g. Thesaurus). Specifically, it deals
with the problem of retrieving similar distributed
words to the newly added one. It uses, in particular,
theorem called additive axiom to calculate the
proximity between two words. This means finding
the nearest word to the added one in order to choose
the suitable mapping schema, and thus, reusing it to
conceive the new word mapping. Let’s present this
additive axiom in what follows. Let the precision
P
add
be the result returned from the calculation of the
proximity between two words: w and w’, with taking
into account the common features between these two
words, P
add
є [0, 1]. This precision is calculated
according to the following formula:
P
ad
d
(w, w’) = ∑
TP
(
w
,
w’
)
D
t
yp
e
(w, c)
(1)
∑
F
(
w
)
D
t
yp
e
(w, c)
The weight D determines which word occurrence is
important enough to be presented in its description.
There are various weight functions, e.g.:
1 if P(c|w) > 0,
(2)
D
t
yp
e
(w, c) = 0 otherwise.
With P(c|w) the probability of the w occurrence.
Let F(w) presents the set of all the properties of a
word w. F(w) = {c: D
type
(w, c) > 0}, with D the
weight associated to w and c the occurrence number
(word appearance number in the results returned by
the already executed users’ queries).
The TP(w, w’) corresponds to the shared properties
between two words: w and w’ (TP(w, w’) = F(w) ∩
F(w’)). The precision P
add
allows to find w: the
nearest word to the newly added word w’. It
calculates the proximity between this w’ and each
word w
i
of existing words. The calculation of this
proximity is based on two basic principles: (i) the
common features between two words and (ii) the
importance of the old word. The next section shows
how these parameters are insufficient for an
effective retrieval of the nearest word.
3 PRECISION AND SEMANTICS
IN ADDITIVE AXIOM
The calculation of the word importance in the
additive axiom introduced in (Weeds and Weir,
2005) is relaying on some parameters like
occurrence number in executed query’s results. But,
since the number of useless word occurrence is
sometimes important, the word importance
calculation using only this occurrence number
parameter turns out insufficient. To solve this
problem, it is necessary to introduce other
calculation parameters. Thus, we propose to add two
other parameters (i) the similarity calculation
(degree) between two words (the added word and an
existing one) and (ii) the consideration of the user’s
opinion note (user’s satisfaction degree) associated
to every word with respect to the old returned users’
query results.
The similarity between two words w and w’ is
calculated by measuring the distance between them
d
w
(w, w’) (Zhong et al., 2002). To calculate this
distance, we consider the place (position) of words
with respect to its root ccp (Zhong et al., 2002):
d
w
(w, w’) = d
w
(w, ccp) + d
w
(w’, ccp) with d
w
(w,
ccp) = milestone(ccp) – milestone(w) and milestone
is the word position value: milestone(n) = 1 / 2kl(n)
(with k the speed by which the value decreases along
the hierarchy and l(n) indicates the depth of the word
in the hierarchy (e.g. l(cpp) = 0)). Then, the
similarity calculation formula is: Sim
w
(w, w’) = 1 –
d
w
(w, w’); with 0 < Sim
w
< 1. This parameter can
add more accuracy at the selection of the nearest
word to the added one.
Besides, for the users’ opinion note parameter,
old queries’ results should be used to benefit from
the domain experts’ knowledge. So, the integration
system has to allow the user introducing his opinion.
Therefore, each user notes his satisfaction degree
related to each word received in his queries’ results.
EFFICIENT SEMI-AUTOMATIC MAINTENANCE OF MAPPING BETWEEN ONTOLOGIES IN A BIOMEDICAL
ENVIRONMENT
259
The reuse of these results can help to improve the
importance word calculation by returning a more
significant selection results. The value of a user’s
opinion note for a given word is the average of all
users’ opinions related to this word (the sum of all
users’ opinions values divided by users’ number):
S
w
= Avg((S
w
)i), 0 < i < n; with n the number of
users. This S
w
value is always included between 0
and 1 (0 < S
w
< 1) since the precision P
add
can not
exceed 1. Hence, after a user’s query execution, a
value is associated to every word in this query which
is added precisely to the description of the word.
Then, after the insertion of these two new
parameters, the new axiom formula will be:
P
add
(w, w’) = ∑
TP(w, w’)
D
type
(w, c) * Sim
w
(w, w’) * S
w
∑
F(w )
D
type
(w, c)
(3)
The calculation of “Sim
w
(w, w’) * S
w
” add more
precision to define the nearest word to the added one
in a more semantic way. When we associate this
calculation to the other which calculates the
proximity between two words in initial P
add
, we can
conclude that our solution brings a more accurate
answer in the selection of the nearest word to the
added one.
In the following section, we show through an
illustrative example that these two added parameters
allow a better reliability. We also show that the use
of the initial additive axiom is not sufficient in the
case of important occurrence number of useless
words. So, we allow reusing the mapping of the
nearest element found in a reliable way.
4 ILLUSTRATIVE EXAMPLE
We adopt the additive axiom presented in (Weeds
and Weir, 2005) and add modifications (addition of
similarity calculation and the user’s opinion note) to
adapt it to our requirements. Indeed, the axiom
which we have just defined calculates the precision
of every added word with respect to each existing
word. We have to look for the nearest word to the
added one by defining the most suitable additive
model (model generated by the additive axiom for
the word and relation additions between them). We
note this as: Max {P
add
(w
i
, w’)} with i all existing
words.
We explain the initial additive axiom limit as
well as the improvements that it will take through an
illustrative example.
4.1 Application Example of Sim
w
and
S
w
Parameters
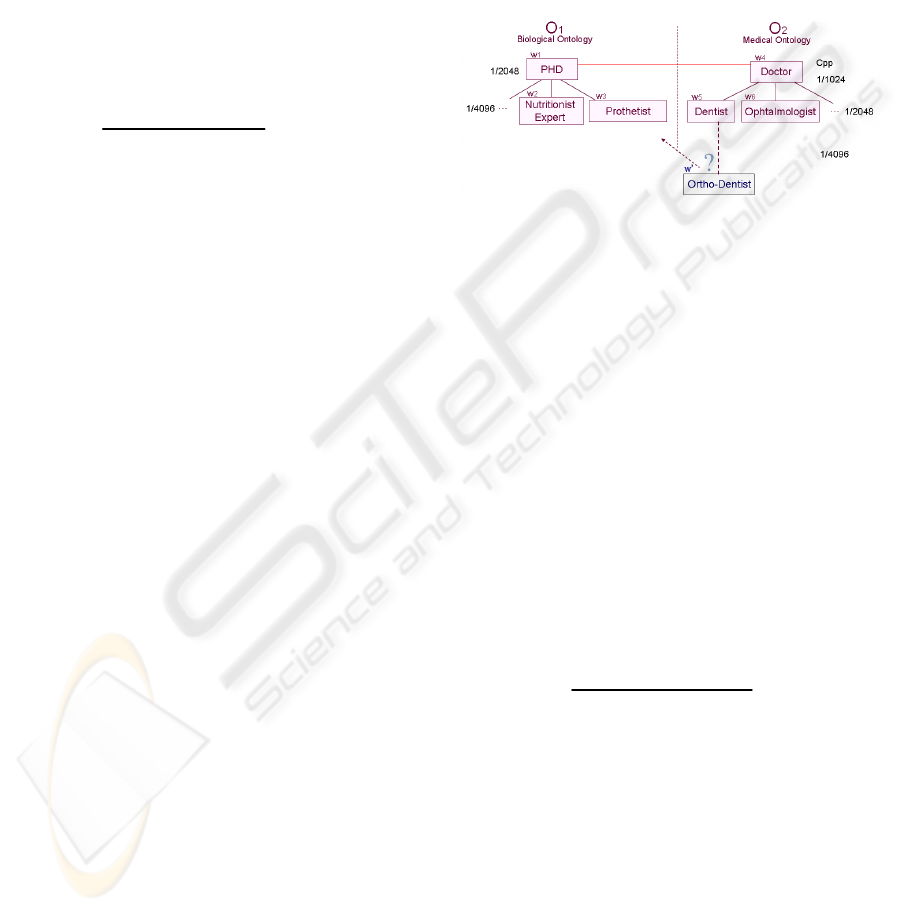
Let O
1
be a first ontology of biological domain and
O
2
a second one of medical domain.
Let w’: ‘Ortho-Dentist’ be the newly added word
and w
i
be the set of existing words in both
ontologies O
1
and O
2
: w
1
: ‘PHD’, w
2
: ‘Nutritionist
Expert’, w
3
: ‘Prosthetiste’, w
4
: ‘Doctor’, w
5
:
‘Dentist’, w
6
: ‘Ophtalmologist’.
Figure 1: Word Addition.
Finding the nearest word to ‘Ortho-Dentist’
means calculating the proximity between w’
(‘Ortho-Dentist’) and every word w
i
by P
add
precision formula.
First, let’s apply the initial axiom: (1).
So, let’s begin by calculating the proximity
between the added word ‘Ortho-Dentist’ (w’) and a
word among the existing words such as ‘Prosthetist’
(w
3
).
D
type
(w
3
, c) = 1, represents the weight associated
with the word w
3
since the number of its occurrence
can be important.
Let ∑F(w
3
) = 10, be the w
3
features (attributes)
and ∑F(w’) = 7, be the w’ features.
TP(w
3
, w’) = F(w) ∩ F(w’), ∑TP(w
3
, w’) = 4,
which represents the common features between w
3
and w’.
Then the precision calculated would be:
P
add
(w
3
, w’) = ∑
TP(w3, w’)
D
type
(w
3
, c) = 0.40000.
∑
F(w3)
D
type
(w
3
, c)
By the same way we obtain also the following
precisions for the other words:
P
add
(w1, w’) = 0.10041, P
add
(w5, w’) = 0.39999 and
P
add
(w4, w’) = 0.21551.
Now, let’s apply the new axiom (extended) by
taking into account both added parameters.
We begin by looking for the nearest word to
‘Ortho-Dentist’. Let’s take the example of the word
‘Doctor’ (w
1
) and let’s calculate P
add
(w
1
, w’). We
begin by calculating the distance as we mentioned
before:
d
w
(Ortho-Dentist, Doctor) = d
w
(w
1
, w’)
= d
w
(w
1
, w
4
) + d
w
(w’, w
4
)
ICEIS 2010 - 12th International Conference on Enterprise Information Systems
260

= (1 / 1024 – 1 / 4096) + (1 / 1024 – 1 / 2048)
= 0.00073.
Then similarity will be:
Sim
w
(w
1
, w’) = 1 – 0.00073 = 0.99878.
The similarity calculation of these three other
words (w
5
: ‘Dentist’, w
4
: ‘Physician’ and w
3
:
‘Prosthetist’) is measured by the same manner as w
1
,
so we obtain: Sim
w
(w
5
, w’) = 0.99878, Sim
w
(w
4
, w’)
= 0.99927 and Sim
w
(w
3
, w’) = 0.99854.
Let S
w1
= 0.7, S
w5
= 0.6, S
w4
= 0.5 and S
w3
= 0.6.
The S
w
values are chosen little close but different
and not too small voluntarily to highlight the
influence of the user’s opinion in our P
add
precision
calculation. This calculation does not focus only on
the user’s opinion, but also on the similarity
calculation which we added and the calculation of
common features between the two words already
presented in the initial axiom. So, the values of S
w
should not be also very big to avoid hiding the effect
of these two other parameters.
We can note that S
w5
and S
w3
have the same value
which is chosen voluntarily to show the influence of
the similarity in our calculation.
At last let’s apply the totality of the new axiom:
P
add
(w
1
, w’) = 0.10041*0.99878*0.7 = 0.07020,
P
add
(w
5
, w’) = 0.39999*0.99876*0.6 = 0.23969,
P
add
(w
4
, w’) = 0.21551*0.99926*0.5 = 0.10767 and
P
add
(w
3
, w’) = 0.40000*0.99853*0.6 = 0.23964.
4.2 Comparison between Initial and
New Axiom Results
In the following, we detail the values obtained for
each word with respect to the newly added word.
We mention the values obtained by the initial
additive axiom and those obtained by adding the two
new parameters. We get so the following table:
Table 1: Comparison of axioms’ results.
Initial Axiom’s
P
add
New Axiom’s
P
add
PHD (i = 1) 0.10041 0.07020
Dentist (i = 5) 0.39999
0.23969
Doctor (i = 4) 0.21551 0.10767
Prosthetist (i = 3)
0.40000
0.23964
Using the initial additive axiom, the nearest word
to w’ (‘Ortho-Dentist’) is w
3
(‘Prosthetist’) which
corresponds to the maximal value of P
add
precision
(Max(P
add
(w
i
, w’)) = P
add
(w
3
, w’)). Whereas, the
introduction of similarity and user’s opinion
parameters makes w
5
(‘Dentist’) appears as the
nearest word to w’ (Max(P
add
(w
i
, w’)) = P
add
(w
5
,
w’)). This last result is closer to the reality, since
‘Ortho-Dentist’ is a speciality of ‘Dentist’ and they
are both doctors, which is not the case of
‘Prosthetist’. In other words, contrary to the initial
axiom, the implementation of the extended axiom
formula allows to realize that the word ‘Prosthetist’
does not be the semantically nearest word to the
added one ‘Ortho-Dentist’ although its occurrence
number is important in the users’ query results.
Indeed, the calculation of “Sim
w
(w, w’)” adds
more precision to define w as the nearest word to w’.
The calculation of S
w
refines the selection of the
word w in a semantic way, by using the old queries’
results corresponding to each word w.
The mapping schemas of w are selected to reuse
them in creating the w’ mapping schema.
Consequently, the additive model of w’ is created in
an efficient way.
5 RELATED WORK
Several research works focus more and more on the
schema mapping thanks to the big interest accorded
to the integration and so to the mapping schema. The
mapping can decline into three classes (Choi et al.,
2006): (i) mapping between domain ontology and
data source local ontologies, (ii) mapping between
local ontologies and (iii) mapping for ontology
merging.
Managing this mapping in an automatic way
constitutes a real challenge because of the enormous
quantity of biomedical data sources. As it is
explained earlier, a semi-automatic mapping can
reduce the users’ effort (Aumueller et al., 2005). The
already existing mapping schemas can be shared and
reused to avoid recreation tasks, e.g. QuickMig
(Drumm et al., 2007) which uses new techniques for
the exploitation of the existing mappings’ schemas
by detecting not only the correspondence elements
but also the mapping’s expressions. The example of
COMA (Do and Rahm, 2002) is also based on the
principle of old mapping reuse, combining several
techniques of mapping. This system was extended in
order to support schemas and ontologies written with
various format types, what gave birth to COMA++
(Aumueller et al., 2005). This last system adopts a
new mapping technique for ontologies and reduces
the execution time. To choose the most suitable
mapping schema for the added element, the nearest
element to the newly added one has to be selected.
Then, its mapping schema can be reused (Madhavan
EFFICIENT SEMI-AUTOMATIC MAINTENANCE OF MAPPING BETWEEN ONTOLOGIES IN A BIOMEDICAL
ENVIRONMENT
261

et al., 2001, Do and Rahm, 2002 and Weeds and
Weir, 2005). This last study uses the similarity and
word importance calculation between words to
choose the nearest element.
6 CONCLUSIONS
In this paper we have dealt with the retrieval of the
nearest element to an added one in order to reuse its
mappings. Therefore, we have extended an existing
additive axiom formula by introducing new
parameters for the effective retrieval of the nearest
element (word) to the added one. First, we introduce
the similarity calculation between the added word
and existing ones. This enables more precision and
accuracy in the calculation of the semantic proximity
between two words. Second, we take into account
the users’ opinions to measure the importance of a
word with respect to its semantic value. This allows
emphasizing the semantic importance of concepts
(words) with respect to experts’ opinion. The
introduction of these two parameters allows
covering more semantic heterogeneity between data
sources of the biomedical domain. In consequence, it
allows an efficient semi-automatic mapping
maintenance. Such mapping can be very useful for
diverse research studies which are interested in the
integration of heterogeneous data sources distributed
on large scale. We validate our method by an
illustrative example of the extended additive axiom
including the proposed two parameters. We show
that results obtained by our method are closer to the
reality than those found by the initial axiom.
For future works, we are planning to design a
better evaluation of our contribution by relying on
real experiments. We can also test the extended
additive axiom when thousand of heterogeneous
types of data are added to diverse domain
ontologies.
REFERENCES
Aumueller, David, Do, Hong-Hai, Massmann, Sabine and
Rahm, Erhard, 2005. Schema and Ontology Matching
with COMA++. ACM SIGMOD International
Conference on Management of Data, Baltimore,
Maryland.
Choi, Namyoum, Song, Il-Yeol and Han, Hyoil, 2006. A
Survey on Ontology Mapping. ACM SIGMOD
Record.
Couto, Francisco M., Silva, Mario J., Coutinho and Pedro
M., 2007. Measuring semantic similarity between
Gene Ontology terms. Data & Knowledge
Engineering Journal.
Davidson, S., Overton, C. and Buneman, P., 1995.
Challenges in Integrating Biological Data Sources.
Journal of Computational Biology.
Do, Hong-Hai and Rahm, Erhard, 2002. COMA-A system
for flexible combination of schema matching
approaches. VLDB Conference, Hong Kong, China.
Doan, AnHai, Madhavan, Jayant, Domingos, Pedro and
Halevey, Alon, 2002. “Learning to Map between
Ontologies on the semantic Web”. WWW Conference,
Honolulu, Hawaii, USA.
Drumm, Christian, Schmitt, Matthias and Do, Hong-Hai,
2007. QuickMig-Automatic Schema Matching for
Data Migration Projects. ACM CIKM Conference,
Lisbon, Portugal.
Hakimpour, Farshad and Geppert, Andeas, 2002. Global
Schema Generation Using Formal Ontologies.
International Conference on Conceptual Modeling,
Finland.
Hammer, J. and Schneider, M., 2003. Genomics Algebra:
A New, Integrating Data Model, Language, and Tool
Processing and Querying Genomic Information. CIDR
Conference, Asilomar, CA.
Hernandez, Thomas and Kambhampati, Subbarao, 2004.
Integration of Biological Sources: Current Systems
and Challenges Ahead. ACM SIGMOD Record.
Jonquet, Clement, Musen, Mark A. and Shah, Nigam,
2008. A System for Ontology-Based Annotation of
Biomedical Data. DILS Workshop, Paris, France.
Karmakar, Samir, 2007. Designing Domain Ontology: A
Study in Lexical Semantics. Technical Report, Indian
Institute of Technology Kanpur, Kanpur, India.
Madhavan, Jayant, A. Bernstein, Philip and Rahm, Erhard,
2001. Generic Schema Matching with Cupid.
Microsoft Research (extended version of VLDB
Conference paper).
Melnik, Sergey, Garcia-Molina, Hector and Rahm, Erhard,
2002. Similarity Flooding: A Versatile Graph
Matching Algorithm and its Application to Schema
Matching. ICDE Conference, San Jose, CA.
Silva, Nuno and Rocha, Joa, 2003. “MAFRA-An
Ontology Mapping FRAmework for the Semantic
Web”.
International Conference on Business
Information Systems, USA.
Weeds, Julie and Weir, David, 2005. Co-occurrence
Retrieval: A Flexible Framework for Lexical
Distributional Similarity. MIT Press Journals.
Zhong, Jiwei, Zhu, Haiping, Li, Jianming and Yu, Yong,
2002. Conceptual Graph Matching for Semantic
Search. ICCS Conference, Borovets, Bulgaria.
ICEIS 2010 - 12th International Conference on Enterprise Information Systems
262
