
An Image-based Framework for Intellectual Decision
Making in Medical Research
Sara Colantonio
1
, Igor Gurevich
2
, Ovidio Salvetti
1
and Yulia Trusova
1
1
Institute of Information Science and Technologies, Italian National Research Council
Via G. Moruzzi 1, 56124 Pisa, Italy
2
Institution of the Russian Academy of Sciences “Dorodnicyn Computing Center of RAS”
Vavilov str. 40, 119333 Moscow, Russian Federation
Abstract. Medical decisional problems strongly rely on the analysis and inter-
pretation of diagnostic images acquired by different investigation techniques.
Information extraction and correlation from these are usually demanding tasks
that can burden the routine work of clinicians. In this paper, an image-based
framework is presented which is devoted to the automated extraction of know-
ledge and data from biomedical images used for intellectual decision making in
clinical and medical research. An ontological approach is adopted for encoding
different kind of knowledge required in problem solving. Modelling the pre-
clinical stage of Parkinson's disease is presented as eligible case study.
1 Introduction
An important step in medical diagnostics is analysis of data extracted from digital
images, i.e. microscopic, X-ray, MRT and ultrasonic images. For efficient assistance
of problem solving is necessary to automate of biomedical image analysis and recog-
nition and to develop suitable information technologies and tools.
During the last years, authors’ research has been focused on the investigation and
development of image mining methods for solving and improving knowledge in
medical tasks, in particular, for disease modelling and diagnostics. In previous works
[1, 2], a novel knowledge-based approach to medical image mining was described.
The approach is based on an ontological representation of the knowledge needed for
problem solving. Ontologies are intended to describe the following kinds of knowl-
edge: 1) knowledge on image processing, analysis, storage, and retrieval; 2) knowl-
edge on processing and analysis of experimental data; 3) knowledge on prediction of
diseases progression and treatment results. The proposed approach was developed as
a general framework for medical image mining, and then specialized for automation
of cytological image analysis used for early diagnostics of oncological blood diseases.
In particular, the cell image analysis ontology was developed.
The general value of the framework is allowing the development of a new appli-
cation devoted to investigate the definition of a preclinical model of Parkinson’s
disease (PD). More precisely, thanks to the flexible design of the framework, the new
Colantonio S., Gurevich I., Salvetti O. and Trusova Y. (2010).
An Image-based Framework for Intellectual Decision Making in Medical Research.
In Proceedings of the Third International Workshop on Image Mining Theory and Applications, pages 60-67
DOI: 10.5220/0002962500600067
Copyright
c
SciTePress

application is just requiring the definition of a specific part of the ontological knowl-
edge base, i.e., the domain ontology and base of rules related to the PD medical do-
main.
In the next sections, the framework is briefly described, while attention is focused
on the specific problem domain and the dedicated knowledge base that is being de-
veloped.
2 Related Works
Ontologies as an effective way for knowledge representation became very popular
last years. Different works related to usage of ontologies for solving image-based
tasks have been reported. For example, in [3] an approach for solving the symbol
grounding problem involved in semantic image interpretation is presented. The
method is based on using the image processing ontology to reduce the gap between
the image processing level and the visual level. Authors note that the proposed ontol-
ogy is not complete and should be considered as a basis for further extension. In [4] a
platform dedicated to the knowledge extraction and management for image process-
ing applications is proposed. It includes a system that automatically generates image
processing applications on the basis of goal formulations given by a user who is inex-
perienced in image processing domain. The user defines the goal of processing in
terms of his/her application domain and then the system translates this information
into image processing terms taken from the image processing ontology. The result of
this translation is an image processing request which is sent to the planning system to
generate the program that responds to this request.
The main contribution of our work is the development of a sufficiently detailed
and well-structured ontology which will cover all important aspects of image process-
ing, analysis and understanding (main categories of concepts, their properties and
relations). The proposed ontology can be used as a base for the construction of spe-
cialized knowledge bases for supporting image analysis and, then, image mining.
3 The Knowledge-based Framework for Image Mining
One of actual and difficult problems in modern medicine is the development of sys-
tems for early diseases diagnostics at the preclinical stage. The automation of diag-
nostics requires automated extraction of information from medical images used for
diseases modelling, treatment and intellectual decision making.
To this end, a knowledge-based framework was conceived for aiding the investi-
gation and diagnostic processes of medical problems, by providing local and remote
access to known cases, the facility for retrieving images by content, and the possibili-
ty of mining image patterns relevant to medical decision making, e.g., diagnoses and
prognoses [1].
Several practical scenarios can be supplied for highlighting the usefulness of
framework functionalities:
61
− Clinicians’ case-based reasoning, i.e., medical decisions made by retrieving simi-
lar cases according to patient’s information on imagery data;
− Investigation of concurrent situations, i.e., exploration of other research results of
the same kind but in other fields;
− Extraction of new knowledge, i.e., investigation on the relevance of new informa-
tion (new data, new parameters, new relations among them) for some still debated
problems.
A brief overview of the framework can be useful to recall and present its main
functionalities and component.
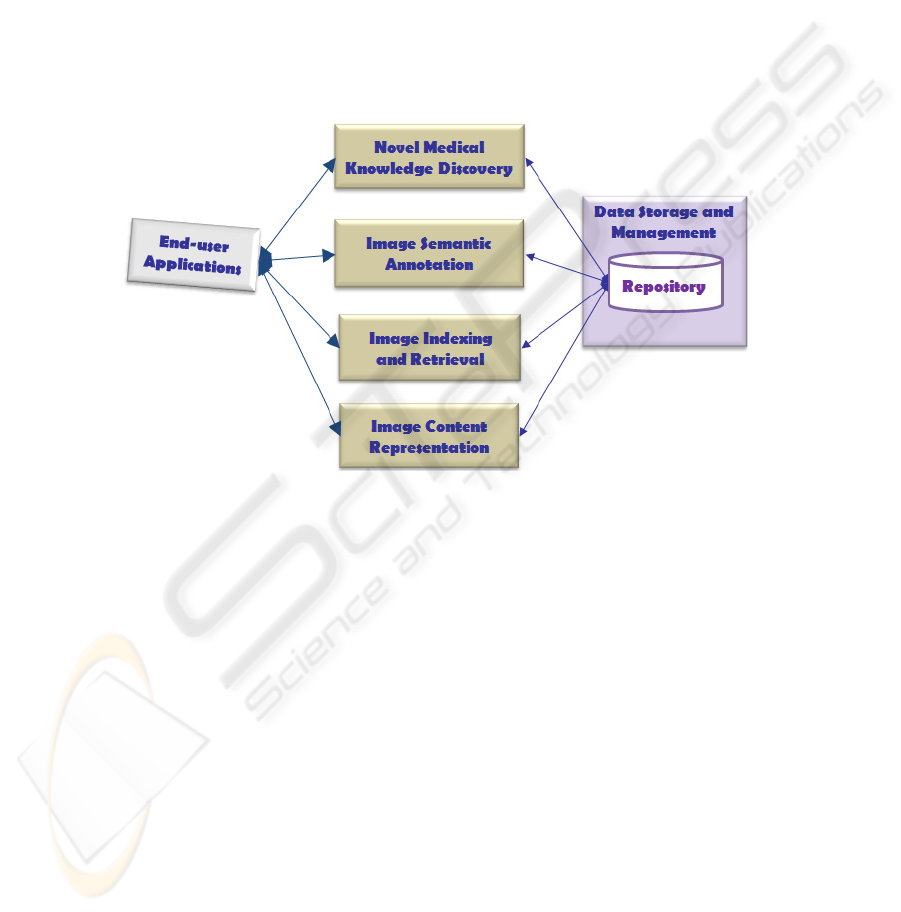
The framework was functionally designed as shown in Fig. 1. More precisely,
along with the data storage and management inside a repository, three main and inter-
related end-users functionalities were identified:
Fig. 1. The knowledge-based framework functionalities.
Image indexing and retrieval, for finding images ranked in accordance to some
requirements on their content. Retrieval can be performed by means of explicit
text query or by supplying a reference image. Similarity measures are applied to
appropriate image representations for identifying the relevant images to be re-
trieved;
Image semantic annotation, for defining a list of semantic keywords to be asso-
ciated to image and used for their retrieval, in particular for text queries. A struc-
tured terminology is supplied for aiding annotations by the users;
Novel medical knowledge discovery, for extracting valid, novel and understanda-
ble knowledge about the diagnostic, prognostic and monitoring processes. Ad-
vanced data mining methods can be applied to patterns built by correlating fea-
tures extracted from images and domain concepts.
The main components of framework, as sketched in Figure 2, are:
a repository for storing, accessing and retrieving images and information extracted
at different levels from them;
62
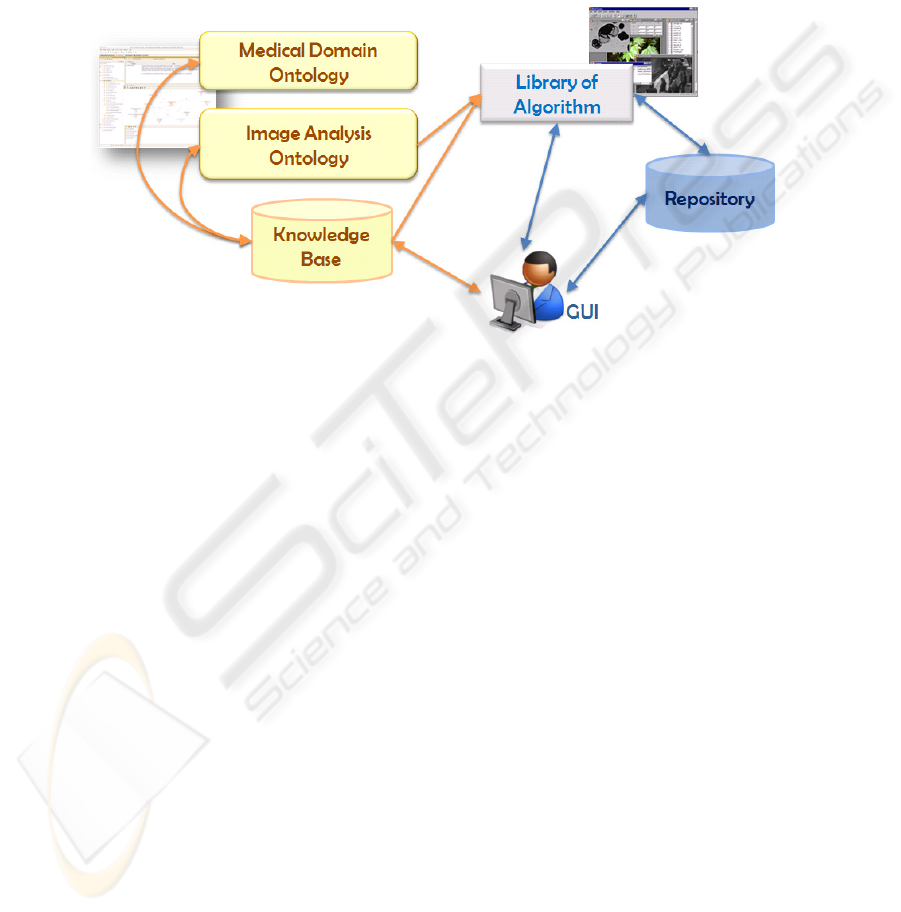
a suite of ontologies including specific domain ontologies related to medical prob-
lems and a general image analysis ontology [7];
a collection of algorithms for image processing, analysis, recognition and mining;
a user interface for accessing, uploading, browsing and annotating images.
For a detailed description of these components see [1], here we concentrate on the
specific application and, hence, on the definition of the Brain Image Mining Ontolo-
gy.
Fig. 2. The main framework components.
4 Diagnosis of Neurodegenerative Diseases
The framework is being extended in order to suitably define a model of preclinical
stage of Parkinson's disease. In the next subsections, the problem domain and the
framework extensions are presented.
4.1 Automated Early Diagnostics of Neurodegenerative Diseases
The investigation and modelling of PD is a crucial and always present problem in
medicine. Construction of experimental models is crucial for the research of neurode-
generative disease pathogenesis. PD is characterized by a progressive degeneration of
dopaminergic (DA-ergic) neurons in the substantia nigra pars compacta (SN) leading
to a dopamine (DA) depletion in the striatum. As a result, parkinsonian patients lose
the ability to control their movements.
Current tasks are automation of experimental data extraction for filling a model of
PD preclinical stage and automation of model investigation by means of computer
experiments.
The development of PD preclinical stage model is a complex screening analysis
which is being done cooperatively by physiologists, biochemists and morphologists.
63

The morphological research requires processing and analysis of astronomical quantity
of experimental animal serial brain section images. And studying of each of the sec-
tions requires quantitative and qualitative feature measurements and analysis of sev-
eral thousand neurons and axons [5].
The initial data is digital images of the immunostained sections of various brain
areas. DA-ergic neurons were labelled on serial sections (a thickness of 20 microns)
of the substantia nigra (Fig. 3) and their fibres (axons) on sections of the striatum (a
thickness of 12 microns) (Fig. 4) by immunohystochemestry for tyrosinehydroxylase
(TH) (TH is the specific enzyme of DA synthesis). Experimental data has been re-
ceived from digital images of distal parts of axons (terminals).
Fig. 3. Neurons. Fig. 4. Terminals.
Terminals are small rounded objects with an area varying from 0.6 to 3 m2. Ter-
minals can have an oval, round, prolate or irregular shape. In the presented gray-scale
images, the brightness of terminals is lower than the background brightness.
The PD model represents the differences between experimental and control
groups. The former is a group of animals injected with a toxin, while the latter is a
group of animals not affected by the toxin. A major characteristic of the PD model is
the number of DA-ergic axons innervating the striatum in the case of using various
schemes for neurotoxin administration (dose, the number of injections, intervals be-
tween injections). The extent of degeneration is defined as the difference between the
number of terminals of DA-ergic axons in the control and experimental groups. DA-
ergic neurons and axons remaining after neurotoxin administration are supposed to
demonstrate increased functional activity in order to compensate for the DA defi-
ciency. An indicator of the increased functional activity of neurons and their fibres
can be an increase in their sizes. An increase in the concentration of tyrosinehydroxy-
lase (key enzyme in DA synthesis) is supposed to be another specific indicator of the
functional activity of DA-ergic axons and neurons.
4.2 The Brain Image Knowledge Base
The study of PD can be performed from different perspectives which should be con-
sidered as a whole to have a better view of the disease. More specifically, in the sys-
tem physiology perspective, the structures (anatomy) involved in the disease are con-
64

sidered, while the ways those structures interact or fail to interact belong to the physi-
ology perspective of the disease state. Moreover, the genetic perspective is becoming
more and more practised, since the discovery that genetic mutations in the alpha
synuclein gene could cause PD has opened new avenues of research in PD.
The idea behind the extension of the knowledge based framework is to provide
PD researchers with an instrument that allows them to search for particular patients
and particular images of disease states, correlate different kinds of data (of the same
patient or of different patients), to formulate hypotheses on the disease state, causes
and evolution. Examples of usage of the framework can be the following [6]:
A researcher interested in anatomy/physiology trying to hypothesize the location
of various components on a neuron might want to look at scientific data or hy-
potheses on the presence of certain type of receptors on a neuron.
A researcher interested in molecular biology trying to hypothesize the location of
some receptors on neurons might want to look at scientific data or hypotheses on
the anatomical structure of a neuron or the pharmacology of chemical compounds
that bind to a receptor.
A clinical researcher interested in developing new therapies for a disease may be
interested in understanding the mechanisms of how chemical compounds or
ligands bind to receptors.
For providing these functionalities, a knowledge base able to integrate different
kind of data is being developed. In particular, the information required belongs to:
anatomy and physiology;
molecular biology;
clinical information.
Such knowledge base is being developed by integrating a dedicated ontology and
a base of rules which use the ontology concepts and are aimed at encoding more
complex relations among them. Several existing biomedical ontologies have been
taken into account for reuse [6]. Since the idea is to consider all the information be-
longing to the three domains listed above, sub-hierarchies related to neuron anatomy
(class Neuron, and all the meronimy classes), proteins, genes and general information
related to disease are being considered. Figure 5 shows a fragment of the ontology.
The rules are being encoded by discussing with domain experts who suggest some
relevant relations among domain relevant data and information.
Moreover, the library of algorithms is being extended by inserting methods for
image segmentation and classification, according to the specifications contained in
[5].
In this way, the ontology can be used to annotate images and retrieve particular
cases according to users’ requirements. On the other hand, the most complex informa-
tion contained in the knowledge base will allow users to formulate hypotheses and
check their validity but retrieving images and data that support and evidence its valid-
ity.
65
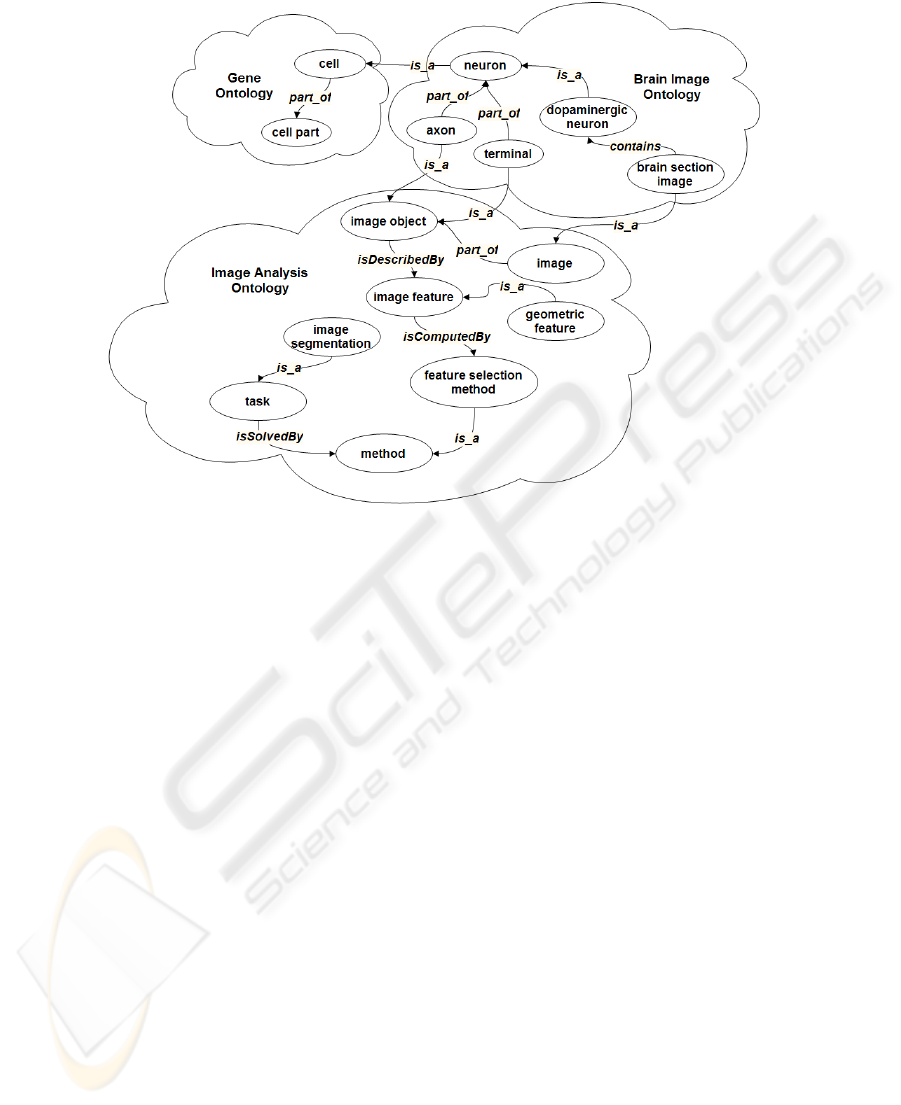
Fig. 5. The fragment of the Brain Image Mining Ontology.
5 Conclusions
An image-based framework to biomedical image mining has been presented. The
main attention was focused on the problem of extraction and representation of knowl-
edge and data required for intellectual decision making in clinical and medical re-
search. Modelling the preclinical stage of Parkinson's disease has been considered as
eligible case study. The requirements to the information represented in the knowledge
base on brain image mining have been described.
Future work will be devoted to further development of the knowledge base de-
scribed, in particular, to the completion of the ontology suite aimed at encoding the
knowledge on storing, retrieving, processing, analysis and mining of biomedical im-
ages.
Acknowledgements
This work was partially supported by the Russian Foundation for Basic Research
(project no. 09-07-13595), by the Program of the Presidium of the RAS “Intelligent
information technologies, mathematical modelling, system analysis and automation”
(project 204), by the Foundation for Assistance to Small Innovative Enterprises (con-
tract № 6956p/9009).
66

References
1. Colantonio, S., Gurevich, I., Pieri, G., Salvetti, O., Trusova, Yu.: Ontology-Based Frame-
work to Image Mining // Image Mining Theory and Applications. Proceedings of the 2nd
International Workshop on Image Mining Theory and Applications - IMTA 2009 (in con-
junction with VISIGRAPP 2009), Lisboa, Portugal, February 2009. Portugal: INSTICC
PRESS (2009) 11-19
2. Colantonio, S., Martinelli, M., Salvetti, O., Gurevich, I.B., Trusova, Y.O.: Cell Image
Analysis Ontology. Pattern Recognition and Image Analysis: Advances in Mathematical
Theory and Applications. 18(2) (2008) 332-341
3. Hudelot, C., Maillot, N., Thonnat, M.: Symbol Grounding for Semantic Image Interpreta-
tion: From Image Data to Semantics. In: 10th IEEE International Conference on Computer
Vision (ICCVW'05) (2005) 1875
4. Renouf, A., Clouard, R., Revenu, M.: A Platform dedicated to knowledge engineering for
the development of image processing applications. In: Proceedings of the ICEIS 2007, Vol.
AIDSS, Funchal, Portugal (2007) 271-276
5. Gurevich, I., Koryabkina, I., Kozina, E., Myagkov, A., Niemann, H., Ugrumov, M., Ya-
shina, V.: Automated Extraction of Data for Construction of Parkinson’s Disease Experi-
mental Model. Pattern Recognition and Information Processing (PRIP’2009): Proceedings
of the 10
th
International Conference (19-21 May, 2009, Minsk, Belarus) (2009) 308-313
6. Kashyap V., Cheung K., Doherty D., Samwald M., Marshall M.S., Luciano J., Stephens S.,
Herman I., Hookway R.. An Application of Ontology-Based Data Integration for Biomedi-
cal Research. In The Semantic Web. Real-World Applications from Industry. Cardoso,
Hepp, Lytras (Eds.). Springer (2008) 97-122
7. Gurevich, I.B., Salvetti, O., Trusova, Yu.O.: Fundamental Concepts and Elements of Image
Analysis Ontology // Pattern Recognition and Image Analysis: Advances in Mathematical
Theory and Applications 19(4) (2009) 603-611
67
