
Personal Identification and Authentication based on
One-lead ECG using Ziv-Merhav Cross Parsing
David Pereira Coutinho
1,3
, Ana L. N. Fred
2,3
and M´ario A. T. Figueiredo
2,3
1
Instituto Superior de Engenharia de Lisboa, Lisboa, Portugal
2
Instituto Superior T´ecnico, Lisboa, Portugal
3
Instituto de Telecomunicac¸ ˜oes, Lisboa, Portugal
Abstract. In this paper, we propose a new data compression based ECG biomet-
ric method for personal identification and authentication. The ECG is an emerg-
ing biometric that does not need liveliness verification. There is strong evidence
that ECG signals contain sufficient discriminative information to allow the iden-
tification of individuals from a large population. Most approaches rely on ECG
data and the fiducia of different parts of the heartbeat waveform. However non-
fiducial approaches have proved recently to be also effective, and have the advan-
tage of not relying critically on the accurate extraction of fiducia. We propose a
non-fiducial method based on the Ziv-Merhav cross parsing algorithm for symbol
sequences (strings). Our method uses a string similarity measure obtained with
a data compression algorithm. We present results on real data, one-lead ECG,
acquired during a concentration task, from 19 healthy individuals, on which our
approach achieves 100% subject identification rate and an average equal error
rate of 1.1% on the authentication task.
1 Introduction
Biometrics deals with identification of individuals based on their physiological or be-
havioral characteristics [1]. Traditional methods of biometric identification, include
those based on physiological characteristics like fingerprints or iris, and those based on
behavioral characteristics like signature or speech. Although some technologies have
gained more acceptance than others, the field of biometrics for access control plays an
important role in the security at airports, industry and corporate workplaces, for exam-
ple. But some technologies lack robustness against falsification. Some may be based
on such characteristics that for a group of people is difficult to acquire or even that
characteristics is missing.
The electrocardiogram (ECG) is an emerging biometric measure which exploits a
physiological feature that exists on every human and there is a strong evidence that the
ECG is sufficiently discriminative to identify individuals from a large population. The
ECG feature allows liveliness detection (intrinsic), personal identification and authen-
tication, and different stress or emotion states detection [2]. The ECG is a behavioral
biometric trait that can be used with other biometric measures [3], as a complementary
feature, for fusion in a multimodal physiological authentication system [4, Ch. 18] and
for continuous authentication where biological signatures are continuously monitored
Pereira Coutinho D., L. N. Fred A. and A. T. Figueiredo M.
Personal Identification and Authentication based on One-lead ECG using Ziv-Merhav Cross Parsing.
DOI: 10.5220/0003030000150024
In Proceedings of the 10th International Workshop on Pattern Recognition in Information Systems (ICEIS 2010), page
ISBN: 978-989-8425-14-0
Copyright
c
2010 by SCITEPRESS – Science and Technology Publications, Lda. All rights reserved
(easily done by using new signal acquisition technologies like the Vital Jacket [5], [6])
in order to guarantee the identity of the operator throughout the whole process [7].
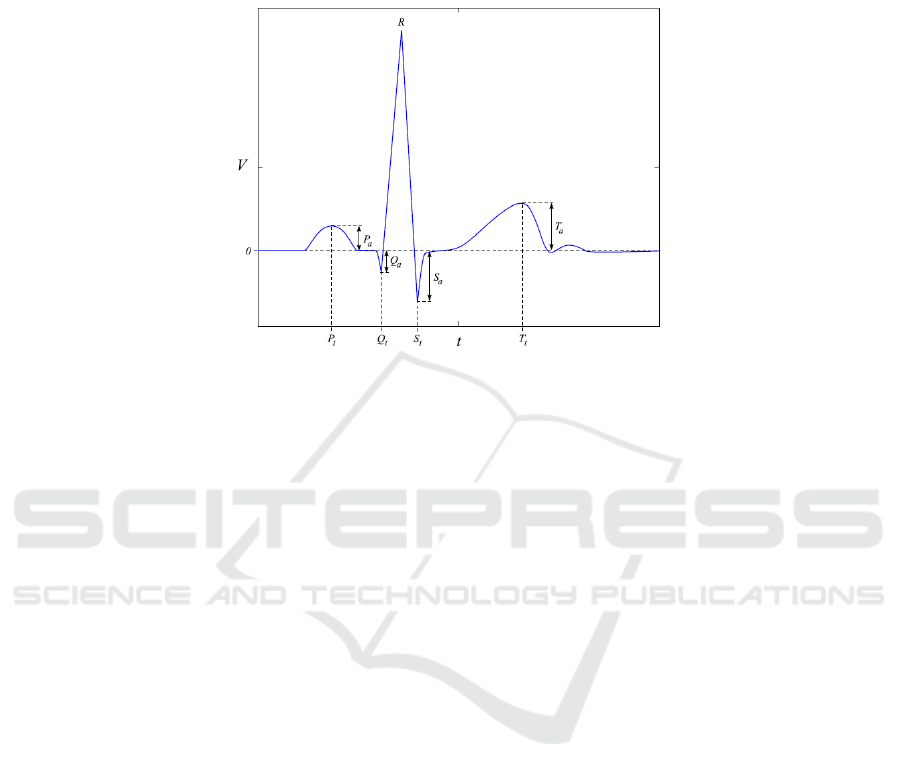
Fig.1. Example of four latency times (features) measured from the P, QRS and T complexes of
an ECG heartbeat for fiducial-based feature extraction.
A typical ECG signal of a normal heartbeat can be divided into 3 parts, as depicted
in Fig. 1: the P wave (or P complex), which indicates the start and end of the atrial
depolarization of the heart; the QRS complex, which corresponds to the ventricular
depolarization; and, finally, the T wave (or T complex), which indicates the ventricular
repolarization. It is known that the shape of these complexes differs from person to
person, a fact which has stimulated the use of the ECG as a biometric [8].
In a broad sense, one can say there are two different approaches in the literature
concerning feature extraction from ECG: fiducial [8], [9], [10], [11], and non-fiducial
[12], [13]. Fiducial methods use points of interest within a single heartbeat waveform,
such as local maxima or minima; these points are used as reference to allow the defi-
nition of latency times, as shown in Fig. 1. Several methods exist that extract different
time and amplitude features, using these reference points. Non-fiducial techniques aim
at extracting discriminative information from the ECG waveform without having to lo-
calize fiducial points. In this case, a global pattern from several heartbeat waveforms
may be used as a feature. Some methods combine these two different approaches or are
partially fiducial [14] (e.g., they use only the R peak as a reference for segmentation of
the heartbeat waveforms).
Biel et al. [8] pioneered the use of the ECG as a biometric for personal identifi-
cation. They used a 12-lead ECG but ended up concluding that one lead was enough
because 12-lead ECG systems require meticulous placement of the electrodes on each
person, which is not practical. Using a proprietary equipment from SIEMENS, 30 fidu-
cial features were extracted; a feature selection algorithm allowed concluding that the
best results were with 10 features. Classification was based on the principal component
analysis (PCA) of each class. The purpose was to identify 20 subjects at rest and they
achieved an accuracy of 100%.
16

Recent studies have shown that non-fiducial approaches also allow successful per-
sonal identification using the ECG heartbeat signal.
Chiu et al. [13], using a one-lead ECG, introduced a system based on a 3-step fea-
ture extraction method. It uses QRS complex detection (with the So and Chan method
[15]) and waveform alignment in the time domain; the features extracted are based on
the discrete wavelet transform. A nearest neighbor classifier based on the Euclidean dis-
tance between pairs of feature vectors is used. The purpose was to identify 35 subjects
(no activity specified) from the QT database [16]. The results obtained were: 100% of
accuracy on person identification and 0.83% FAR (false acceptance rate) and 0.86%
FRR (false rejection rate) for authentication.
This paper introduces a new non-fiducial ECG-biometric method that uses averaged
single heartbeat waveforms and is based on data compression techniques, namely the
Ziv-Merhav cross parsing (ZMCP) algorithm for sequences of symbols. We present
results on real data, using one-lead ECG acquisition during a concentration task. Notice
that a study [2] with the dataset showed the existence of differentiated states in the
data representing the ECG signal of a subject due to detectable changes along the time
in the acquired signal. On a set of 19 healthy individuals, our method achieves 100%
subject identification (recognition) rate and an average equal error rate of 1.1% on the
authentication (verification) task.
The outline of the paper is as follows. In Section 2, we review the fundamental
tools underlying our approach: Lempel-Ziv string parsing and compression; the Ziv-
Merhav cross parsing algorithm. Section 3 presents the proposed classification method.
Experimental results are presented in Section 4, while Section 5 concludes the paper.
2 The Lempel-Ziv and Ziv-Merhav Algorithms
The Lempel-Ziv (LZ) algorithm is a well-known tool for text compression [17], [18],
[19], [20], which in recent years has also been used for classification purposes (see [21]
and references therein). In particular, in [21], we have shown how the Ziv-Merhav (ZM)
method for measuring relative entropy [22] (which is based on Lempel-Ziv-type string
parsing) achieves state-of-the-art performance in a specific text classification task. We
will now briefly review these algorithms.
– The incremental LZ parsing algorithm [18], is a self parsing procedure of a se-
quence into c(z) distinct phrases such that each phrase is the shortest sequence that
is not a previously parsed phrase. For example, let n = 11 and z = (01111000110 ),
then the self incremental parsing yields (0, 1, 11, 10, 00, 110), namely, c(z) = 6.
– The ZM (cross parsing) algorithm, a variant of the LZ parsing algorithm, is a se-
quential parsing of a sequence z with respect to another sequence x (cross parsing).
Let c(z|x) denote the number of phrases in z with respect to x. For example, let
z be as above and x = (10010100110); then, parsing z with respect to x yields
(011, 110, 0 0110), that is c(z|x) = 3.
Roughly speaking, we can see c(z) as a measure of the complexity of the sequence
z, while c(z|x), the code-length obtained when coding z using a model for x (cross
parsing), can be seen as an estimate of the cross complexity [23]. It is expectable that
17
the cross complexity is low when the two sequences are very similar; this is the key
idea behind the use of ZM cross parsing in classification [21], which in this paper will
be adopted for ECG-based personal identification and authentication.
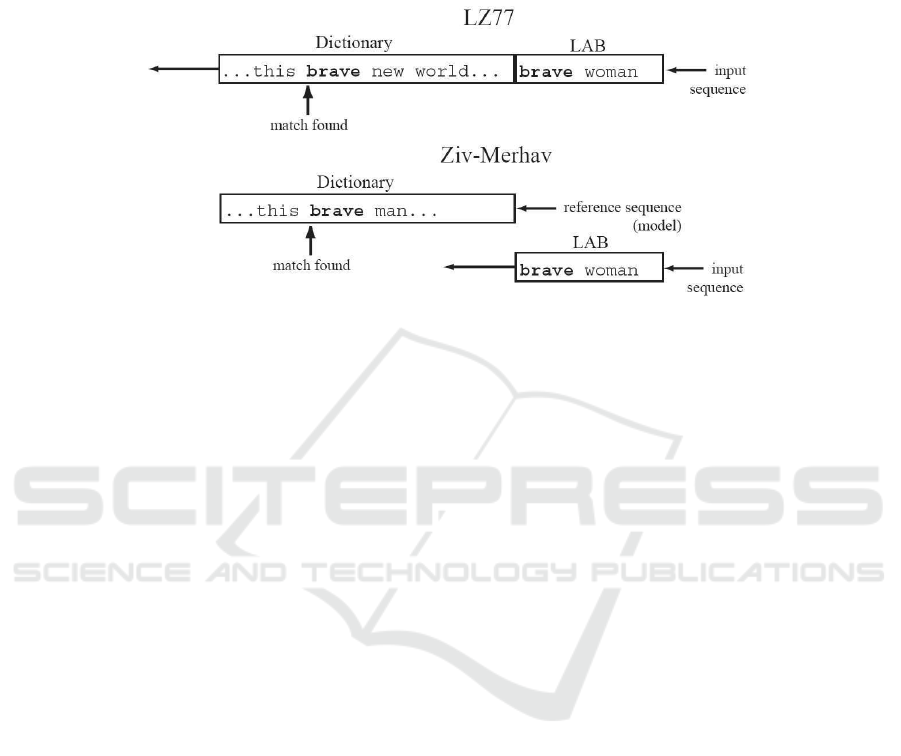
Fig.2. The original LZ77 algorithm uses a sliding window over the input sequence to update the
dictionary; in our implementation of ZM cross parsing algorithm, the dictionary is static and only
the lookahead buffer (LAB) slides over the input sequence.
An implementation of the ZM cross parsing algorithm as a component of a ZM
method for relative entropy estimation was proposed in [21], based on a modified LZ77
[17] algorithm, where the dictionary is static and only the lookahead buffer slides over
the input sequence, as shown in Fig. 2 (for more details, see [21]). This very same
implementation of the cross parsing, using a 64 Kbyte dictionary and a 256 byte look
ahead buffer, was used in the experiments reported below.
3 Proposed Methods
To use ZM-based tools for identification or authentication, a necessary first step is the
conversion of the ECG (discrete-time analog) signal into a sequence of symbols (text).
In this paper, we propose a very simple approach based on quantization. Assuming we
are given a set of single heartbeat waveforms (resulting from a segmentation prepro-
cessing stage), we simply apply 8-bit (256 levels) uniform quantization, thus obtaining
a sequence of symbols (from a 256 symbols alphabet) from each single heartbeat.
Quantizers with fewer bits were considered in early experiments but discarded be-
cause they didn’t perform as well as the 8-bit quantizer. Higher values were not consid-
ered for sake of system implementation simplicity and because of the good performance
obtained with 8 bits.
Consider a collection of training samples partitioned into K classes (the set of sub-
jects to be identified): X = {X
1
, X
2
,..., X
K
}. For each subject/class k, X
k
contains n
strings obtained from the same number of heartbeats using the quantization procedure
described in the previous paragraph. A string x
k
is formed by concatenating the n train-
ing strings of subject k; string x
k
is, in some sense, a “model” representing the shape
of the heartbeats of subject k.
18

3.1 Identification
Given a test sample z (containing the string representing m heartbeats) obtained from
an unknown subject (assumed to be one from which the training set was obtained), its
identity is estimated as follows:
ˆ
k(z) = arg min
k∈{1,...,K}
c(z|x
k
),
where c(z|x
k
) is computed by the ZM cross parsing (ZMCP) algorithm, as described
in Section 2. In other words, the test sample is classified as belonging to the subject that
leads to its shortest description. Although using different tools, this approach is related
in spirit with the minimum incremental coding length (MICL) approach [24].
3.2 Authentication
The authentication (verification) procedure depends on a threshold level, which depends
itself from the range of values of c(z|x
k
). In order to limit its variation to a predefined
set of values, normalization is used. Since in the worst case the description length, re-
sulting from the ZMCP algorithm for the test sample z, is the length of z, the normalized
description length c
n
(z|x
k
) is defined as follows:
c
n
(z|x
k
) =
c(z|x
k
)
len(z)
,
where len(z) is the number of bytes in test sample z. Notice that c
n
(z|x
k
) ∈ [0, 1].
Test sample verification is made by comparing the value of c
n
(z|x
k
) when using the
claimed identity model with a threshold value ∈ [0, 1], previously set according to a
selected error rate, false acceptance rate (FAR), or false rejection rate (FRR). It decides
for genuine when the comparison result is less or equal to the selected threshold level.
4 Experiments
The architecture of the proposed ECG-based biometric system for person identification
and authentication follows the same model proposed by Jain et al in [1]. Fig. 3 shows
the block diagram of the implemented system for the authentication task.
19

one template
User interface
Feature Extractor Matcher (1 match)
System
Database
Preprocessing Quantization ZMCP Comparator
Name (PIN)
True/False
threshold
Fig.3. Block diagram of the implemented system, for the authentication task, is shown using the
five main modules of a biometric system, i.e., sensor, preprocessing, feature extraction, matcher
and system database.
4.1 Data Collection
The ECG waveform dataset used was acquired using one lead, in the context of the
Himotion project.
4
The dataset contains ECG recordings from 19 subjects acquired
during a concentration task on a computer, designed for an average completion time of
10 minutes. All the acquired ECG signals were normalized and band-pass filtered (2–
30Hz) in order to remove noise. Each heartbeat waveform was sequentially segmented
from the full recording and then all the obtained waveforms were aligned by their R
peaks. From the resulting collection of ECG heartbeat waveforms, the mean wave for
groups of 10 consecutive waveforms (without overlap) was computed. Each of these
mean waveforms is what we call a single heartbeat in Section 3.
An intra-class study [2] with the dataset, in the context of the exploration of elec-
trophysiological signals for emotional states detection, showed the existence of differ-
entiated states in the data that represent the ECG signal of a subject. To deal with this
intra-class differences the proposed method includes in the “model” (as mentioned in
Section 3) single heartbeats randomly selected from the whole ECG signal sample.
The reported results are averages over 50 runs. In each run, we partition the set of
heartbeats of each subject into two mutually exclusive subsets: one of these subsets is
used to form the training data set X = {X
1
, X
2
,..., X
K
}, while the other is used to build
the test waveforms. We consider several values for n (the length of the “model” strings)
as well as for m (the length of the test waveforms).
4.2 Identification Results
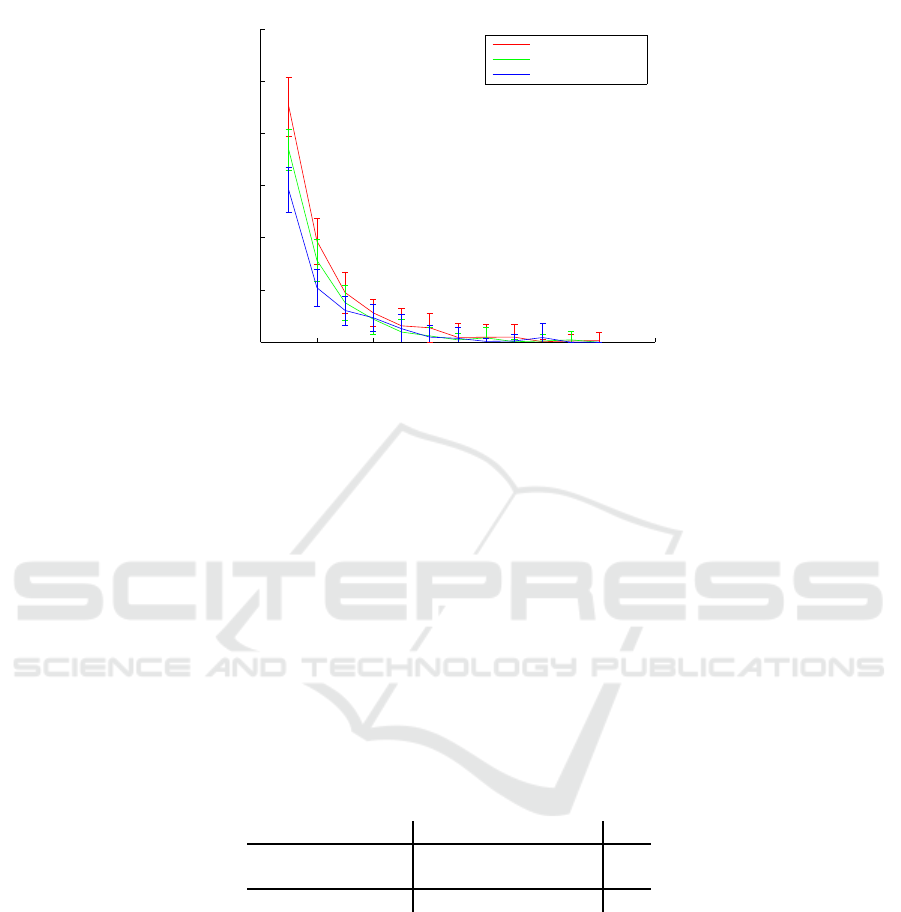
The results for the identification experiment, which are depicted in Fig. 4, show that the
proposed method achieves 100% accuracy for m = 12 and n = 13 or n = 20. This
is better than the results reported in [11] over the same dataset. The approaches in [8],
[9], [13], were not tested on the same dataset, so the results are not directly comparable.
Notice that using only m = 5 waveforms for the test patterns, we already reach an
accuracy around 99.5%. As expected, the accuracy increases both with n and m.
4
https://www.it.pt/auto temp web page preview.asp?id=305
20
0 2 4 6 8 10 12 14
0
2
4
6
8
10
12
Number of test waveforms
Error [%]
10 waveforms model
13 waveforms model
20 waveforms model
Fig.4. Mean recognition error and standard deviation intervals for subject identification when
considering a variable number of waveforms as test samples.
4.3 Authentication Results
Regarding verification (authentication) three different test were made. The first test fol-
lows the model shown in Fig. 3. The results, which are depicted in Fig. 5 (a), show
that the proposed method achieves an overall equal error rate EER ≈ 6%. Notice that
one can lower the error rate using lower threshold values but then the system will reject
more legitmate users. However, it is possible to use lower threshold values if we use a
different value for each subject (user-tuned thresholds).
The second test also follows the model shownin Fig. 3 but now the threshold is user-
tuned. An equal error rate (EER) was computed for each subject and then an average
EER is reported. The test results presented in Table 1 show that the proposed method
outperforms fiducial approaches results reported in [25] and [26], over the same dataset.
Table 1. Comparison of verification related work results with our method, over the same dataset.
Reference Feature EER
Oliveira and Fred [26] Fiducial (1-NN classifier) 8.0 %
Gamboa [25] Fiducial (user tuned) 1.7 %
Proposed method Non-fiducial (user tuned) 1.1 %
On the last verification test, we evaluate the combination of multiple source acqui-
sition signals, classified by a bank of classifiers with the same structure of the first test,
shown in Fig. 3, and a final decision made according to the majority voting criterion.
Given a test sample (of length m = 12), it was decomposed in 64 different ways into
samples of length m = 6 which were classified by a bank of 64 classifiers using the
same threshold level and the same database. The results in Fig. 5 (b) show that this
multiple classifier strategy doesn’t improve the performance.
21
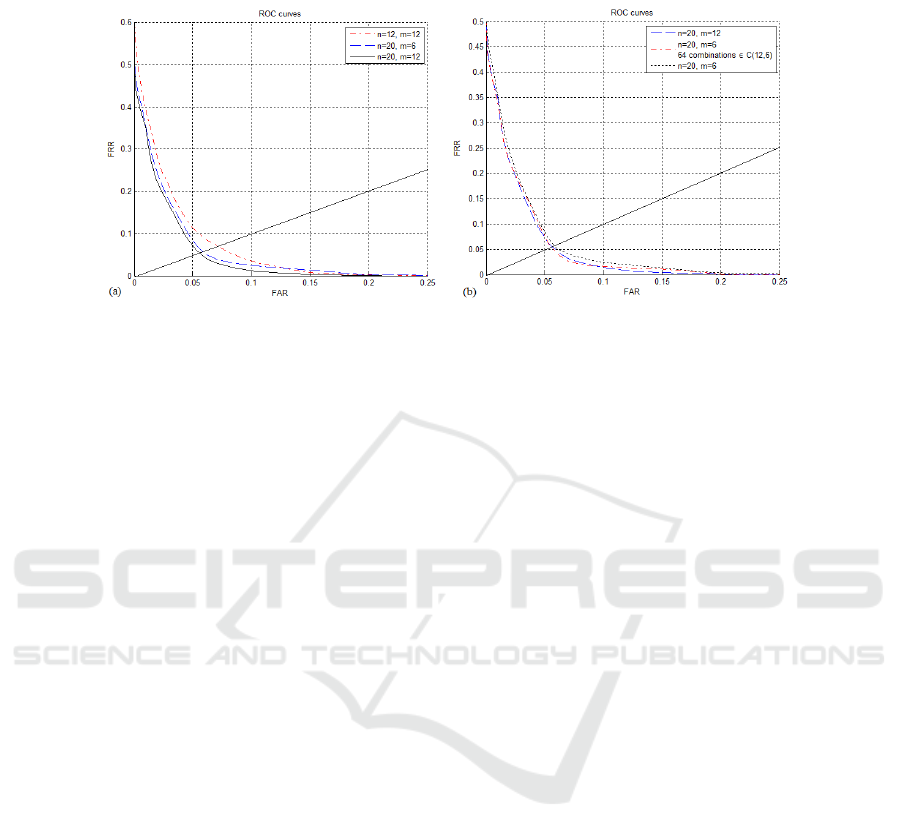
Fig.5. ROC curves for the verification task (the solid straight line has slope 1, for reference pur-
poses). Left plot: results for different n and m values; notice the improvement with the increase
of m and n. The best equal error rate (EER) is close to 6%. Right plot: results for single classi-
fiers versus a bank of 64 classifiers with the same structure for combination of multiple source
acquisition signals.
5 Summary and Conclusions
We have presented a method for personal identification and authentication from one-
lead ECG signals which involves no explicit feature extraction other than 8 bit uniform
quantization of the waveforms. The classifier is based on the Ziv-Merhav cross pars-
ing (ZMCP) algorithm, which is an estimator of the algorithmic cross-complexity [23],
used to measure the similarity between the model waveforms and the test waveforms.
Experiments carried out on a dataset with 19 healthy subjects, for whom the existence
of differentiated states in the ECG data of a subject has been shown [2], showed that
our method achieves100% accuracy in recognition (identification) and an average equal
error rate close to 1.1% in verification (authentication) tasks. Although further experi-
ments, on other datasets, are needed to assess the relative performance of the proposed
method, with respect to other state-of-the-art techniques, these results demonstrated the
validity of our approach as a tool for personal identification and authentication, and
of the ECG signal as a viable biometric. Future work will include tests with the Max-
Lloyd quantizer and further evaluation of our method when used in an adaptive way for
authentication purposes with continuous biometrics systems [7].
Acknowledgements
We acknowledge the following financial support: the FET programme, within the EU
FP7, under the SIMBAD project (contract 213250);Fundac¸˜ao para a Ciˆencia e Tecnolo-
gia (FCT), under grants PTDC/EEA-TEL/72572/2006 and QREN 3475; Departamento
de Engenharia de Electr´onica e Telecomunicac¸˜oes e de Computadores, Instituto Supe-
rior de Engenharia de Lisboa.
22

References
1. Jain, A.K., Ross, A., Prabhakar, S.: An introduction to biometric recognition. IEEE Trans.
Circuits Syst. Video Techn. 14 (2004) 4–20
2. Medina, L.A.S., Fred, A.L.N.: Genetic algorithm for clustering temporal data - application
to the detection of stress from ecg signals. In: Proc 2nd International Conference on Agents
and Artificial Intelligence (ICAART). (2010) 135–142
3. Ross, A.A., Nandakumar, K., Jain, A.K.: Handbook of Multibiometrics (International Series
on Biometrics). Springer-Verlag New York, Inc., Secaucus, NJ, USA (2006)
4. N. V. Boulgouris (Editor), Konstantinos N. Plataniotis (Editor), E.M.T.E.: Biometrics: The-
ory, Methods, and Applications. Wiley-IEEE Press (2009)
5. Cunha, J., Cunha, B., Xavier, W., Ferreira, N., Pereira, A.: Vital-jacket: A wearable wireless
vital signs monitor for patients mobility. In: Proceedings of the Avantex Symposium. (2007)
6. Leonov, V.: Wireless body-powered electrocardiography shirt. In: Proceedings of the Smart
Systems Integration European Conference. (2009)
7. Damousis, I.G., Tzovaras, D., Bekiaris, E.: Unobtrusive multimodal biometric authentica-
tion: the humabio project concept. EURASIP J. Adv. Signal Process 2008 (2008) 1–11
8. Biel, L., Pettersson, O., Philipson, L., Wide, P.: ECG analysis – a new approach in human
identification. IEEE Transactions on Instrumentation and Measurement 50 (2001) 808–812
9. Shen, T., Tompkins, W.J., Hu, Y.H.: One-lead ECG for identity verification. Proc. of the 2nd
Joint Conf. of the IEEE Eng. in Medicine and Biology Soc. and the 24th Annual Conf. and
the Annual Fall Meeting of the Biomedical Eng. Soc. (EMBS/BMES ’02) 1 (October 2002)
62–63
10. Israel, S.A., Irvine, J.M., Cheng, A., Wiederhold, M.D., Wiederhold, B.K.: ECG to identify
individuals. Pattern Recognition 38 (2005) 133–142
11. Silva, H., Gamboa, H., Fred, A.: One lead ECG based personal identification with feature
subspace ensembles. In: MLDM ’07: Proceedings of the 5th international conference on
Machine Learning and Data Mining in Pattern Recognition, Berlin, Heidelberg, Springer-
Verlag (2007) 770–783
12. Chan, A., Hamdy, M., Badre, A., Badee, V.: Wavelet distance measure for person identifica-
tion using electrocardiograms. Instrumentation and Measurement, IEEE Transactions on 57
(2008) 248–253
13. Chiu, C.C., Chuang, C.M., Hsu, C.Y.: A novel personal identity verification approach using
a discrete wavelet transform of the ECG signal. In: MUE ’08: Proceedings of the 2008 In-
ternational Conference on Multimedia and Ubiquitous Engineering, Washington, DC, USA,
IEEE Computer Society (2008) 201–206
14. Wang, Y., Agrafioti, F., Hatzinakos, D., Plataniotis, K.N.: Analysis of human electrocardio-
gram for biometric recognition. EURASIP J. Adv. Signal Process 2008 (2008) 19
15. So, H., K.L.Chan: Development of qrs detection method for real-time ambulatory cardiac
monitor. IEEE Engineering in Medicine and Biology Society. (1997) 289–292
16. Laguna, P., Mark, R., Goldberg, A., Moody, G.: A database for evaluation of algorithms for
measurement of qt and other waveform intervals in the ECG. In: Computers in Cardiology
1997. (1997) 673–676
17. Ziv, J., Lempel, A.: A universal algorithm for sequential data compression. IEEE Transac-
tions on Information Theory 23 (1977) 337–343
18. Ziv, J., Lempel, A.: Compression of individual sequences via variable-rate coding. IEEE
Transactions on Information Theory 24 (1978) 530–536
19. Nelson, M., Gailly, J.: The Data Compression Book. M&T Books, New York (1995)
20. Salomon, D.: Data Compression. The complete reference. 3rd edition edn. Springer, New
York (2004)
23

21. Pereira Coutinho, D., Figueiredo, M.: Information theoretic text classification using the
Ziv-Merhav method. 2nd Iberian Conference on Pattern Recognition and Image Analysis –
IbPRIA’2005 (2005)
22. Ziv, J., Merhav, N.: A measure of relative entropy between individual sequences with ap-
plication to universal classification. IEEE Transactions on Information Theory 39 (1993)
1270–1279
23. Cerra, D., Datcu, M.: Algorithmic cross-complexity and relative complexity. In: DCC ’09:
Proceedings of the 2009 Data Compression Conference, Washington, DC, USA, IEEE Com-
puter Society (2009) 342–351
24. Wright, J., Ma, Y., Tao, Y., Lin, Z., Shum, H.: Classification via minimum incremental
coding length. SIAM Journal on Imaging Sciences 2 (2009) 367–395
25. Gamboa, H.: Multi-Modal Behavioral Biometrics Based on HCI and Electrophysiology. PhD
thesis, Instituto Superior T´ecnico, Universidade T´ecnica de Lisboa, Lisboa, Portugal (2008)
26. Oliveira, C., Fred, A.L.N.: ECG-based authentication: Bayesian vs. nearest neighbour clas-
sifiers. In: Proc International Conf. on Bio-inspired Systems and Signal Processing - Biosig-
nals - INSTICC, Porto, Portugal (2009)
24
