WEIGHTED TIME WARPING FOR TEMPORAL SEGMENTATION
OF MULTI-PARAMETER PHYSIOLOGICAL SIGNALS
Gartheeban Ganeshapillai and John Guttag
Massachusetts Institute of Technology, CSAIL, 77 Massachusetts Avenue, Cambridge, MA, U.S.A.
Keywords:
Temporal segmentation, Multi-parameter physiological signals, Time warping, Time series.
Abstract:
We present a novel approach to segmenting a quasiperiodic multi-parameter physiological signal in the pres-
ence of noise and transient corruption. We use Weighted Time Warping (WTW), to combine the partially
correlated signals. We then use the relationship between the channels and the repetitive morphology of the
time series to partition it into quasiperiodic units by matching it against a constantly evolving template. The
method can accurately segment a multi-parameter signal, even when all the individual channels are so cor-
rupted that they cannot be individually segmented.
Experiments carried out on MIMIC, a multi-parameter physiological dataset recorded on ICU patients, demon-
strate the effectiveness of the method. Our method performs as well as a widely used QRS detector on clean
raw data, and outperforms it on corrupted data. Under additive noise at SNR 0 dB the average errors were
5.81 ms for our method and 303.48 ms for the QRS detector. Under transient corruption they were 2.89 ms
and 387.32 ms respectively.
1 INTRODUCTION
In this paper, we address the problem of segmenting a
quasiperiodic multi-parameter physiological signal in
the presence of noise and transient corruption.
Early warning systems in an ICU require contin-
uous uninterrupted real-time monitoring of the phys-
iological signals (Tarassenko et al., 2006; Mark and
Shavdia, 2007; Cao et al., 2008; Henriques and
Rocha, 2009; Chen et al., 2009). Unfortunately, these
signals often suffer transient corruption. An algo-
rithm that estimates the correct values of the corrupted
signals can help automated systems produce more re-
liable results, and make them more amenable for vi-
sual inspection. An accurate segmentation makes the
estimation task easier.
We represent a multi-parameter signal by a ma-
trix S
n×m
, where each column represents an individ-
ual channel of the signal (e.g., an ECG channel, ABP
or PPG) and each row represents a point in time. For
simplicity, we assume that all the channels are sam-
pled at the same rate. If the matrix represents a quasi-
periodic multi-parameter signal (QPMS), it will have
a repetitive structure that is shared by all the channels
in the structure. This is common in situations where
the signal is generated by the same underlying system.
In the case of the cardiovascular system, the periodi-
1000 1200 1400 1600 1800 2000 2200
−5000
−4000
−3000
−2000
−1000
0
1000
2000
3000
4000
Sample number
Magnitdue
An excerpt from MIMIC dataset
ECG
ABP
Plethysmogram
C
B
A
Figure 1: The Electrocardiogram (ECG), Arterial Blood
Pressure (ABP) and the Photo Plethysmogram (PPG) ex-
tracted from MIMIC, record 039.
city of all the ECG channels, the blood pressure wave-
forms, and the plethysmogram are related to the heart
rate.
In practice, samples may be corrupted in an un-
known fashion. We would like to estimate the actual
values of the samples in row i of the matrix S that
represents a QPMS, using the corrupted values in that
row and the estimates of prior values up to row (i −1).
125
Ganeshapillai G. and Guttag J..
WEIGHTED TIME WARPING FOR TEMPORAL SEGMENTATION OF MULTI-PARAMETER PHYSIOLOGICAL SIGNALS.
DOI: 10.5220/0003128501250131
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2011), pages 125-131
ISBN: 978-989-8425-35-5
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

However, many physiological signals (e.g., those
related to cardiovascular activity) are quasiperiodic.
For such signals, the estimation process starts by iden-
tifying the segment boundaries.
This paper presents a novel approach to identi-
fying segment boundaries in the presence of signifi-
cant amounts of transient corruption spanning multi-
ple columns and rows of the matrix S. The key idea
is that by simultaneously considering all the channels
one can segment them more accurately than would be
possible by considering each channel independently.
The method is based on matching a sliding window
of the QPMS to a template. The template is a short
multi-parameter signal that is regularly updated based
upon recent estimates. The initial template is derived
from an archived QPMS.
Segmentation is done by finding the prefix of
the window that most closely matches the template.
The matching is done using a new method, Weighted
Time Warping (WTW) that minimizes the weighted
morphological dissimilarity between template and the
prefix of the window across all the channels. The
morphological dissimilarity is given by the warped
distance between a channel in the window and the
corresponding channel in the template. The weight
represents the estimated quality of the channel.
Experiments carried out on MIMIC (Goldberger
et al., 2000), a publicly available multi-parameter
physiological dataset recorded from ICU patients,
demonstrate the effectiveness of the method. We con-
sidered additive noise and the following types of cor-
ruptions: signal interruption, exponential damping,
overshooting, clipping, and superimposition of artifi-
cial low frequency signals and high frequency signals.
Here, the corruptions across the channels are not per-
fectly correlated.
We compared our method to a widely used QRS
detection based segmentation method. Our method
performs as well as the QRS detector on the raw data
and significantly outperforms it on the data syntheti-
cally altered with additive noise and transient corrup-
tion. When additive noise at SNR 0 dB was applied to
all m channels of the signal, the average errors were
5.81 ms for our method and 303.48 ms for the QRS
detector. The average errors were 2.89 ms and 387.32
ms respectively when transient corruption was added.
In the last two cases, the QRS detector was totally
unusable, whereas our method was able to do the seg-
mentation with reasonable accuracy.
The primary contributions of this work are
• Formulation of the problem of segmentation as a
dissimilarity minimization problem, and the use
of a new dissimilarity metric, weighted time warp-
ing.
• The use of a multi-parameter signal template that
eliminates the need of any prior knowledge of the
specific properties of the signal, and
• The dynamic adaptation of the template, which al-
lows us to accommodate the time evolution of the
signal.
2 BACKGROUND AND RELATED
WORK
Physiological signals in the ICU are often severely
corrupted; corruptions by noise, artifact and missing
data lead to serious errors in automated medical sys-
tems and early warning systems. (Li et al., 2008) pro-
vides a survey of strategies used to address this prob-
lem. Recent attempts to mitigate this problem focus
on using redundant measurements, and fusing data
from multiple sensors (Li et al., 2008; Aboukhalil
et al., 2008; Li and Clifford, 2008; Deshmane, 2009).
Typically they employ independent methods on dif-
ferent channels and combine the results only at the
final stage. In contrast, we process all channels si-
multaneously.
Dynamic Time Warping (DTW) is increasingly
being used in temporal segmentation problems (Ko-
var and Gleicher, 2004; Zhou et al., 2008). (Park and
Glass, 2006) uses DTW to segment speakers in an au-
dio signal. DTW has also been used in locating mo-
tion clips (Gleicher and Kovar, 2004) and temporal
segmentation of human motions (Zhou et al., 2008).
(Vlachos et al., 2006) provides an overview on the
implementation of multidimensional Dynamic Time
Warping (DTW) over the L
p
norm. Based on this
work, we propose Weighted Time Warping (WTW), a
novel method that uses time warping to perform seg-
mentation.
WTW generalizes multidimensional DTW by
weighing the individual signals by the signal qual-
ity. In WTW, we also apply the local continuity and
global path constraints that reduce the influence of
outliers. The use of such constraints was proposed
in (Myers et al., 1980).
3 METHOD
First we present the overview of the algorithm, fol-
lowed by the discussion of a single iteration of the
loop. Additional details are provided later in this sec-
tion.
BIOSIGNALS 2011 - International Conference on Bio-inspired Systems and Signal Processing
126

0
50
100
−0.2
0
0.2
0.4
0.6
0.8
1
1.2
−0.6
−0.4
−0.2
0
0.2
0.4
0.6
0.8
1
Time
Relative Amplitude
Relative Amplitude
ABP
ECG
`
`
1
`
2
Figure 2: This template comprises of clean ECG and Ar-
terial Blood Pressure (ABP) waveforms. The positions of
the segment boundaries are denoted by `
1
, `
2
and `. The
template is little longer than two segments. It contains two
full segments of length `
2
− `
1
and ` − `
2
; the length of the
template is `.
3.1 Overview
Goal. Let S ∈ ℜ
n×m
be a multi-parameter time-
series consisting of set of m number of single param-
eter physiological, and Z
`×m
= {Z
j
∈ ℜ
`
}
m
an initial
template.
1
The goal is to segment S into a set of
quasiperiodic units Y = {Y
i
} where Y
i
de f
= S
[p
i
,p
i.+1
)
.
Here, S
[p
i
,p
j
)
denotes the window in the target se-
quence S from time t = p
i
to t = p
j
− 1.
We require the template (Figure 2) to be com-
prised of at least two segments. These segments are
used to find the quasiperiodic unit Y
i
in Y. We also
assume that we know the locations of the segment
boundaries Z.`
1
, Z.`
2
and Z.` in the template. Here,
Z.k denotes the k
th
row in Z. It is a vector of samples
corresponding to time k. The prototypical template is
initially obtained from an archive.
Procedure. We start the process at some arbitrary
point in time p
start
on the signal that is to be seg-
mented. This need not be an actual segment boundary.
We then run the algorithm starting at p
start
, continu-
ously segment S, and add the segments to Y. We also
update Z to reflect the evolution of the time series.
This enables us to accommodate the gradual changes
in the morphology of the signal.
An Iteration. We start the iteration with the extrac-
tion of a window W = S
[p
i
,p
i
+v)
from the time series
1
Bold capital letters represent a matrix S, non-bold cap-
ital letters denote a column vector A, and lower-case letters
denote a scalar w.
data S at p
i
. Here, the window length is given by
v = ` + e, where e is the buffer length. In the follow-
ing discussion, we use j to index the single param-
eter signals. It is preprocessed, and the morpholog-
ical quality estimates {q
j
}
m
are computed, where q
j
represents the morphological similarity between the
channel W
j
from W and the corresponding channel
from the template Z
j
. For each channel j, a pairwise
Euclidean distance matrix pD
j
is calculated between
Z
j
and W
j
(Equation 1-2). The final distance matrix D
is obtained by weighting the pairwise distance matrix
pD
j
with q
j
(Equation 3).
c
x,y
= W
x, j
− T
y, j
(1)
pD
j
=
c
1,1
c
1,2
.. c
1,|Z
j
|
c
2,1
c
2,2
.. c
2,|Z
j
|
... .. .. ..
c
|W
j
|,1
.. .. c
|W
j
|,|Z
j
|
(2)
D =
s
m
∑
j=1
q
j
|pD
j
|
2
(3)
The accumulated distance matrix aD is then com-
puted from D using dynamic programming. For two
single parameter signals A ∈ ℜ
l
a
and B ∈ ℜ
l
b
, the ac-
cumulated distance can be calculated by the following
recursion
aD(A
i
,B
i
) = D(A
i
,B
i
) + min{D(A
i−1
,B
i−1
),
aD(A
i−1
,B
i
),aD(A
i
,B
i−1
)}. (4)
We next check aD for such spurious matchings. When
a spurious matching is encountered, the buffer length
e is increased and the process is repeated. Otherwise
we trim W to obtain W∗ so that it contains only the
portion of the signal that matches the template.
We next use the accumulated distance matrix aD
to find the optimal path alignment between Z and
W∗, as in DTW. From the alignment, we extract the
point W. f ∗ in W∗ that is matched with the segment
boundary Z.`
2
in Z. This corresponds to the segment
boundary we are interested in, because Z.`
2
marks the
end of the first segment in the template. Then, the
corresponding length f ∗ is used to update p
i+1
with
p
i
+ f ∗. A second pass of the algorithm is used to fine
tune the results. For ECG, a peak detector is used in
the second run. Finally S
[p
i
,p
i+1
)
is added to Y. Fol-
lowing the template update, the process is repeated to
find the next segment boundary.
3.2 Weighted Time Warping
We introduce a weighted norm (Equation 3) over the
parameter signals to vary the influence exerted by
each parameter. The morphological quality metric q
j
WEIGHTED TIME WARPING FOR TEMPORAL SEGMENTATION OF MULTI-PARAMETER PHYSIOLOGICAL
SIGNALS
127

captures the morphological similarity between the pa-
rameter signal W
j
and the template Z
j
.
We hypothesize that the corrupted regions of the
signals are morphologically dissimilar to the clean
signals. We estimate the dissimilarity using the
warped distance cd
j
.
cd
j
= min
k
1
k
aD
j
(`,k) (5)
k
j
= argmin
k
1
k
aD
j
(`,k) (6)
Here, aD
j
is the accumulated distance matrix ob-
tained from pD
j
using dynamic programming, k
j
is
the alignment length, and ` is the length of the tem-
plate.
By simply inverting cd
j
we could obtain the mor-
phological quality metric q
j
. Because we use a sin-
gle metric across different channels such as ECG,
ABP and PPG, sometimes two morphologically simi-
lar but time warped sequences from one channel pro-
duce higher cost than two morphologically dissimilar
sequences from another channel. To address this, we
incorporate additional information such as the align-
ment length k
j
and the difference in the standard de-
viations of W
j
and T
j
to obtain q
j
. Shorter alignment
lengths indicate the partial matchings that usually re-
sult in lower costs. Also a significant difference in
the standard deviation between W
j
and T
j
implies that
they represent two dissimilar processes. We also use
a non-linear transformation to amplify the dynamic
range of the values in the following equation that es-
timates the morphological quality.
q
j
= max
0,exp
1 −
λ × cd
j
k
j
×log(std
W
j
+1)
|std
T
j
−std
W
j
|
− 1
(7)
Here, std
T
j
and std
W
j
denote the standard deviation of
T
j
and W
j
respectively. Further, k
j
is the length of the
path alignment corresponding to cd
j
(Equation 5). λ
is a normalization coefficient which was empirically
chosen to be 0.05.
3.3 Path Constraint
In a typical formulation of DTW, the distance func-
tion that is used to solve DTW (Equation 4), allows
any path to be taken from (A
0
,B
0
) to (A
l
a
,B
l
b
). This
makes DTW susceptible to degenerate matchings, es-
pecially in the presence of noise. For example, a long
subsequence of a signal might be matched with a sig-
nificantly shorter subsequence of another signal.
Therefore, we use local continuity and global path
constraints (proposed as Type III and Type IV local
continuity constraints in (Myers et al., 1980)) to pre-
vent such physiologically implausible alignments by
updating Equation 4 with
aD(A
i
,B
i
) = D(A
i
,B
i
) + min{D(A
i−1
,B
i−1
),
aD(A
i−1
,B
i−2
) + D(A
i
,B
i−1
),
aD(A
i−1
,B
i−3
) + D(A
i
,B
i−1
) + D(A
i
,B
i−2
),
aD(A
i−2
,B
i−1
) + D(A
i−1
,B
i
),
aD(A
i−3
,B
i−1
) + D(A
i−1
,B
i
) + D(A
i−2
,B
i
)}.
(8)
This ensures that there are no long horizontal or
vertical paths in the matrix aD along the alignment.
It also results in global path constraints, by excluding
excluding certain parts on the accumulated distance
matrix in which optimal warping paths could lie.
3.4 Templates
The templates (Figure 2) are initially derived from
an archive of the prototypical multi-parameter signal.
They are then updated using the recent signal esti-
mates.
3.4.1 Template Length
When searching for the segment boundaries we only
assume the approximate location of the starting point
(p
i
). This allows us to start the algorithm at an arbi-
trary location (p
start
). It also makes the detection of
the segment boundary p
i+1
less sensitive to an inac-
curacy in determining the previous segment boundary
(p
i
). To accommodate this we use templates that are
more than two segments long.
3.4.2 Template Update
To follow the gradual changes that are common in the
physiological signals, we update the template regu-
larly. However, we do this only when the behavior
is consistent, i.e., the recent estimates of the segment
lengths have gradual variation. In that case, if all the
channels are also clean enough, i.e., q
j
> β; ∀ j, we
update the template by averaging the excerpt of the
last two segments with the time warped version of the
current template (Equation 10). Here, the time warp-
ing is necessary, since the current template Z∗ and the
excerpt of the last two segments Z
0
can be of different
lengths. Finally we normalize the template.
Z
0
= S
[(p
(i−1)
−ε),p
(i+1)
]
;ε ≥ 1 (9)
Z
00
= warp(Z) + ηZ
0
;η ≥ 0 (10)
Z∗ = normalize(Z
00
) (11)
BIOSIGNALS 2011 - International Conference on Bio-inspired Systems and Signal Processing
128

Here, ε ensures that the template consists of at
least two segments. We vary the influence of the re-
cent segments on the template through the constant
η, where η = 0 implies a static template. We chose
η = 0.6 empirically.
3.5 Long Segments
Spurious matchings occur when the segments in the
window W are significantly longer than the segments
in the template Z; therefore the current buffer length
e is not long enough to span at least two segments in
the window. In such cases, the window W = S
[p
i
,p
i
+v)
would not contain two full segments and does not give
the expected matching. In such situations, we have
to repeat the matching with a longer window. We in-
crease v through e and repeat the process with a longer
window. Otherwise we trim the window and obtain
W∗, the portion of the window that matches the tem-
plate.
4 EXPERIMENTAL RESULTS
We applied our method to multi-parameter physiolog-
ical signal data from MIMIC at Physionet.org (Gold-
berger et al., 2000). The database has 72 waveform
records with several annotation sets including ECG
beat labels. It includes recordings from multiple ECG
channels, Arterial Blood Pressure (ABP) and Photo
Plethysmogram (PPG). The signals are sampled at
125 Hz. From this database, we selected 70 records
that contained one continuous hour of at least one
ECG channel, and ABP, PPG or both.
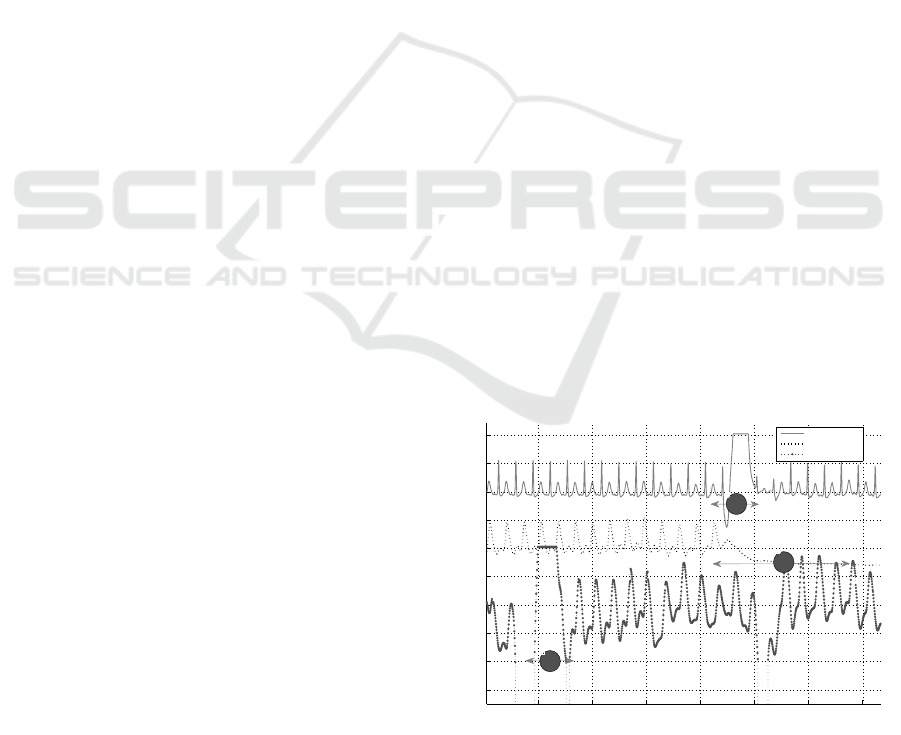
Figure 3: Record 213 contains a) ECG channel I, b) ECG
channel II, c) ECG channel V, d) ABP and e) PPG. The
plethysmogram is completely absent over the 12 hour pe-
riod. A severe corruption spans across all the channels for
significant amount of time also.
We compared our method to a widely used QRS de-
tection based segmentation method (Pan and Tomp-
kins, 1985; Urrusti and Tompkins, 1993). This
method provides high specificity and sensitivity with
low computational load (Kohler et al., 2002). Further
it is publicly available as open source software.
2
We conducted the following experiments : 1) on
the raw MIMIC data we compared our algorithm to
the QRS detector, and 2) we selected clean excerpts
from the raw data, artificially corrupted them, and
compared our algorithm to the QRS detector on this
corrupted data.
We evaluate the performance using the number
of errors and the average error in milliseconds. The
number of errors is defined as the total number of the
estimated segment boundaries that are different from
actual boundaries by more than one sample. This
also includes the cases where a segment boundary
is completely missed and an erroneous boundary is
found. The average error is the mean distance be-
tween an actual segment boundary and the closest
segment boundary found by the method.
4.1 Raw MIMIC Data
We performed two tests on the raw data. In the first
test, we randomly selected 10 records and ran the tests
for the minimum of 12 hours and till the end of the
record. We compared the results of our method and
the QRS detector to the segment boundaries marked
in the data.
The results are presented in Table.1. The results
exhibit significant inter-record deviations. This is
mainly due to the presence of sporadic corruptions in
certain records, which results in a burst of errors in
these records. For example, in record 213 (Figure 3),
the corruption is severe, and it spans all the channels.
It causes significantly large number of errors for both
our method and the QRS detector. The median num-
ber of errors for our algorithm was 65, whereas it was
330 for the QRS detector.
In the second test, we ran the test across 70 records
for an hour. WTW was able to segment 4.121 × 10
5
segments accurately while the QRS detector was able
to segment 4.111 × 10
5
segments correctly. Although
our method made fewer mistakes compared to that
made by the QRS detector, the difference in the accu-
racies of both the methods is not significant because
of the sparsity of the occurrence of corruption in the
raw data.
2
http://www.eplimited.com/
WEIGHTED TIME WARPING FOR TEMPORAL SEGMENTATION OF MULTI-PARAMETER PHYSIOLOGICAL
SIGNALS
129

Table 1: Comparison of WTW against the QRS detector on
selected records.
WTW QRS
Rec avg Sgmt Err avg Err Err avg Err
ID len (ms) # (ms) # (ms)
39 506.25 6 0.01 30 0.25
41 737.91 2 0.00 31 0.47
55 671.13 19 0.05 385 28.27
208 642.67 215 20.63 699 135.39
216 771.33 293 1.10 330 1.64
220 632.88 5 1.65 4 1.20
253 864.40 35 0.15 346 35.13
262 824.93 65 0.30 249 3.55
284 651.70 119 0.23 191 0.96
226 532.17 311 0.55 675 6.25
213 660.31 1921 3.19 2927 42.22
Median number of beats in one record is 56007
4.2 Artificially Corrupted MIMIC Data
4.2.1 Additive Noise
In this experiment we evaluated our algorithm’s per-
formance on noisy data. We obtained clean excerpts
from the raw MIMIC data and corrupted it with artifi-
cial noise. We randomly extracted 1000 high quality
5 minute excerpts for which both our method and the
QRS detector were 100% accurate. We added AWGN
noise at different Signal/Noise (SNR) levels to these
excerpts and tested both the methods on them.
We carried out two tests. In the first test, we added
noise to all m channels, whereas in the second test, we
added noise to only m − 1 channels.
The results of these tests are presented in Table 2.
Our method was able to identify the segment bound-
aries with reasonable accuracy even in the presence
of significant additive noise; further, if any one of the
channel is free of noise, the performance is compara-
ble to the performance on the clean raw data.
4.2.2 Transient Corruption
In this experiment we evaluated our algorithm’s per-
formance on data altered, by transient corruption. We
randomly extracted 1000 high quality 5 minute ex-
cerpts and artificially corrupted them. On each of
these excerpts, we corrupt m − 1 single parameter sig-
nals. For each of these single parameter signals, we
randomly selected 5 non-overlapping 1 minute long
regions and applied one of the following types of cor-
ruptions: signal interruption, exponential damping,
overshooting and clipping, or superimposition of ar-
tificial low frequency signals and high frequency sig-
Table 2: Summary of the experimental results on the artifi-
cially corrupted data.
Average error (ms) on artificially corrupted data
Type QRS WTW
all m any m − 1
AWGN 20 dB 12.87 0.87 0.0097
(SNR) 10 dB 188.11 3.27 0.0011
0 dB 303.48 5.81 0.008
Transient Corruption 387.32 - 2.89
Average segment length is 521 ms.
nals. The ECG channels and either ABP or PPG were
corrupted.
The results are presented in Table 2. Under tran-
sient corruption, the average error was 2.89 ms for
WTW. This is comparable to that of the data with
AWGN at SNR 10 dB on all m parameters. Under
severe transient corruption the QRS detector becomes
totally unusable.
5 CONCLUSIONS
We presented a novel online method for temporal seg-
mentation of quasiperiodic multi-parameter physio-
logical signals in the presence of noise and transient
corruption. Our method uses Weighted Time Warping
to exploit the relationship between the partially corre-
lated channels and the repetitive morphology of the
time series.
Our method has a greater constant overhead in
computational complexity relative to QRS detection
based segmentation algorithm. For a window of
length `, our method uses dynamic time warping
which runs in O(`
2
) time and space. A QRS detection
based segmentation runs in O(`) time. In the exam-
ples used in this paper ` is less than 500.
Our method is particularly useful when the sys-
tem suffers noise and transient corruption. For cor-
ruptions, we tested our method on few artificial cor-
ruptions; but the real world corruptions could be dif-
ferent, and perhaps adversarial. We haven’t tested our
method on these specific types of corruptions. In the
case of additive noise, AWGN is the most difficult to
handle because it spans the entire frequency spectrum
and has the highest uncertainty. Our method performs
well against AWGN.
We chose ABP, PPG and ECG for testing, because
they are commonly available in the recordings of ICU
patients. However, our method should be applicable
to any set of correlated physiological signals such as
Central Venous Pressure (CVP), Pulmonary Arterial
Pressure (PAP) and respiratory signal.
BIOSIGNALS 2011 - International Conference on Bio-inspired Systems and Signal Processing
130

We showed that our method performs as well as
an excellent QRS detector on relatively clean ECG
data. On 1 hour long test data across 70 records, our
method achieved 99.56% accuracy, whereas the QRS
detector achieved 99.41% accuracy. When AWGN is
synthetically added, the difference in the performance
between our method and the QRS detector becomes
significant. Our method was able to limit the aver-
age error to 5.81 ms when all m parameters were cor-
rupted with AWGN at SNR: 0dB, and to 0.008 ms
when m − 1 parameters were corrupted at the same
SNR. The average error for the QRS detector rose to
303.48 ms when the ECG channel is corrupted with
the same AWGN. Similar results were observed on
transient corruption.
REFERENCES
Aboukhalil, A., Nielsen, L., Saeed, M., Mark, R., and Clif-
ford, G. (2008). Reducing false alarm rates for critical
arrhythmias using the arterial blood pressure wave-
form. Journal of biomedical informatics, 41(3):442–
451.
Cao, H., Eshelman, L., Chbat, N., Nielsen, L., Gross, B.,
and Saeed, M. (2008). Predicting icu hemodynamic
instability using continuous multiparameter trends.
Engineering in Medicine and Biology Society, 2008.
EMBS 2008. 30th Annual International Conference of
the IEEE DOI - 10.1109/IEMBS.2008.4650037, pages
3803–3806.
Chen, X., Xu, D., Zhang, G., and Mukkamala, R. (2009).
Forecasting acute hypotensive episodes in intensive
care patients based on a peripheral arterial blood pres-
sure waveform. Computers in Cardiology, 2009 DOI
- UR -, pages 545–548.
Deshmane, A. (2009). False arrhythmia alarm suppression
using ecg, abp, and photoplethysmogram.
Gleicher, M. and Kovar, L. (2004). Automated extraction
and parameterization of motions in large data sets.
ACM transactions on graphics.
Goldberger, A. L., Amaral, L. A. N., Glass, L., Hausdorff,
J. M., Ivanov, P. C., Mark, R. G., Mietus, J. E., Moody,
G. B., Peng, C.-K., and Stanley, H. E. (2000). Phys-
iobank, physiotoolkit, and physionet: Components of
a new research resource for complex physiologic sig-
nals. Circulation, 101(23):e215–e220.
Henriques, J. and Rocha, T. (2009). Prediction of acute
hypotensive episodes using neural network multi-
models. Computers in Cardiology, 36.
Kohler, B.-U., Hennig, C., and Orglmeister, R. (2002).
The principles of software qrs detection. Engineer-
ing in Medicine and Biology Magazine, IEEE DOI -
10.1109/51.993193, 21(1):42–57.
Kovar, L. and Gleicher, M. (2004). Automated extraction
and parameterization of motions in large data sets.
ACM Transactions on Graphics (TOG).
Li, Q. and Clifford, G. D. (2008). Suppress false arrhyth-
mia alarms of icu monitors using heart rate estima-
tion based on combined arterial blood pressure and
ecg analysis. pages 1–3.
Li, Q., Mark, R., and Clifford, G. (2008). Robust heart rate
estimation from multiple asynchronous noisy sources
using signal quality indices and a kalman filter. Phys-
iological measurement.
Mark, R. and Shavdia, D. (2007). Septic shock: provid-
ing early warnings through multivariate logistic re-
gression models.
Myers, C., Rabiner, L., and Rosenberg, A. (1980). Perfor-
mance tradeoffs in dynamic time warping algorithms
for isolated word recognition. Acoustics, Speech
and Signal Processing, IEEE Transactions on DOI -
, 28(6):623– 635.
Pan, J. and Tompkins, W. J. (1985). A real-time qrs detec-
tion algorithm. Biomedical Engineering, IEEE Trans-
actions on DOI - 10.1109/10.725330, BME-32(3):230
– 236.
Park, A. and Glass, J. (2006). A novel dtw-based dis-
tance measure for speaker segmentation. Spoken
Language Technology Workshop, 2006. IEEE DOI -
10.1109/SLT.2006.326807, pages 22–25.
Tarassenko, L., Hann, A., and Young, D. (2006). Integrated
monitoring and analysis for early warning of patient
deterioration. Medical Applications of Signal Process-
ing, 2005. The 3rd IEE International Seminar on (Ref.
No. 2005-1119) DOI -, pages 64–68.
Urrusti, J. and Tompkins, W. (1993). Performance evalua-
tion of an ecg qrs complex detection algorithm. PROC
ANNU CONF ENG MED BIOL.
Vlachos, M., Hadjieleftheriou, M., Gunopulos, D., and
Keogh, E. (2006). Indexing multidimensional time-
series. The VLDB Journal — The International Jour-
nal on Very Large Data Bases, 15(1).
Zhou, F., Torre, F., and Hodgins, J. (2008). Aligned clus-
ter analysis for temporal segmentation of human mo-
tion. Automatic Face & Gesture Recognition, 2008.
FG ’08. 8th IEEE International Conference on DOI -
10.1109/AFGR.2008.4813468, pages 1–7.
WEIGHTED TIME WARPING FOR TEMPORAL SEGMENTATION OF MULTI-PARAMETER PHYSIOLOGICAL
SIGNALS
131
