
BIODBLINK: MULTI-LEVEL DATA MATCHING FOR
AUTOMATIC GENERATION OF CROSS LINKS AMONG
BIOCHEMICAL PATHWAY DATABASES
Jyh-Jong Tsay, Bo-Liang Wu and Hou-Ji Dai
Dept. of Computer Science & Information Engineering, National Chung-Cheng University, Chia-Yi, Taiwan
Keywords: Pathway Database Integration, Data Matching, KEGG, MetaCyc.
Abstract: Most of biological databases provide cross links that point to data records describing the same object in
other databases. However, as more and more databases are available, manually creating and maintaining
cross links becomes very time consuming, if not impossible. Existing databases provide only a small portion
of all possible links. In this paper, we present a database cross link server BioDBLink that can automatically
collect and generate cross links among biological databases. The core of BioDBLink is a data matching
technique that identifies and matches data records or elements describing the same object among pathway
databases. Experiment on a data set collected from several pathway, enzyme and compound databases
shows that our approach is able to identify most of the cross links provided by current databases, discover a
large number of missing links, and detect inconsistency and duplicate errors.
1 INTRODUCTION
Recently, most of the pathway databases provide
cross links to help users to navigate from one
database to another. However, as more and more
databases are available, existing databases often
provide only a small portion of all possible links.
Manually creating and maintaining cross links
becomes very time consuming, tedious and
incomplete. In this paper, we present a software
system BioDBLink that aims to provide a database
link server, and can automatically collect and
generate cross links among biological databases.
The core of BioDBLink is a data matching
technique that identifies and matches data records or
elements describing the same object among pathway
databases. Note that matching of data elements in
pathway databases is by no means easy, and faces
the following sources of problems: errors in
databases, aliases in mames, missing data, and
heterogeneous pathway representations. Errors in
pathway databases are inevitable. A large portion of
pathway information is obtained by literature
curation. There may be errors made in the curated
literatures. In addition, some information is
predicted by computer software. Different databases
may use different software that makes different and
possibly wrong predictions. Synonyms, aliases, and
homonyms are common in biological science. For
example, a compound may have several different
names, and even different formula. Compound
matching simply by names or formula does not work
well. In reaction level, we need to deal with missing
data. For example, some reactions may miss EC-
number or part of compounds. In pathway level, we
need to deal with heterogeneous definitions and
representations of pathways. For example, KEGG
combine the same pathways from different
organisms into the same map. MetaCyc keeps a
separate pathway for each organism.
Several approaches have been proposed to
integrate different types of biological databases
(Birkland and Yona, 2006; Garcia, Chen and Ragan,
2005; Macauley J., Wang, Goodman, 1998;
Krishnamurthy et al., 2003; Chen and Chen, 2006;
Rajasimha, 2004; Tsay, Wu and Chen 2009). Data
matching has played a central role in physical
integration of databases. In this paper, we propose a
multi-level data matching approach that utilizes
three levels of information provided in pathway
databases: compounds, reactions and pathways. We
first use attributes of compounds to identify
matching between compounds, which is then used to
induce matching between reactions. Reaction
125
Tsay J., Wu B. and Dai H..
BIODBLINK: MULTI-LEVEL DATA MATCHING FOR AUTOMATIC GENERATION OF CROSS LINKS AMONG BIOCHEMICAL PATHWAY DATABASES.
DOI: 10.5220/0003136601250130
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2011), pages 125-130
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
matching is used to induce pathway matching as
well as to enhance compound matching.
We experiment our approach on a data set
collected from two well-known pathway databases
KEGG and MetaCyc, and a number of compound
and enzyme databases, including PubChem, ChEBI,
KNApSAcK, LIPIDMAPS, LipidBank, PDB-CCD,
3DMet, NIKKAJI, NCI, and UM-BBD, ExplorEnz,
IUBMB ExPASy and BRENDA. The experiment
shows that our approach is very promising. For
example, between KEGG and MetaCyc, our
approach identifies 6129 pairs of compound
matching, 3608 pairs of reaction matching, and 1483
pairs of pathway relations. According to our
matching result, we assign EC-numbers to 315
reactions in MetaCyc that do not have EC-numbers,
and discover several duplicate errors in both
databases. We use the unification links provided by
MetaCyc to evaluate the matching performance of
our approach. MetaCyc provides 4268 unification
links to KEGG for compounds, and 24 of them are
invalid links. Our approach discovers 4098 of 4244
valid links. The recall with respect to the set of
unification links in MetaCyc is 0.966.
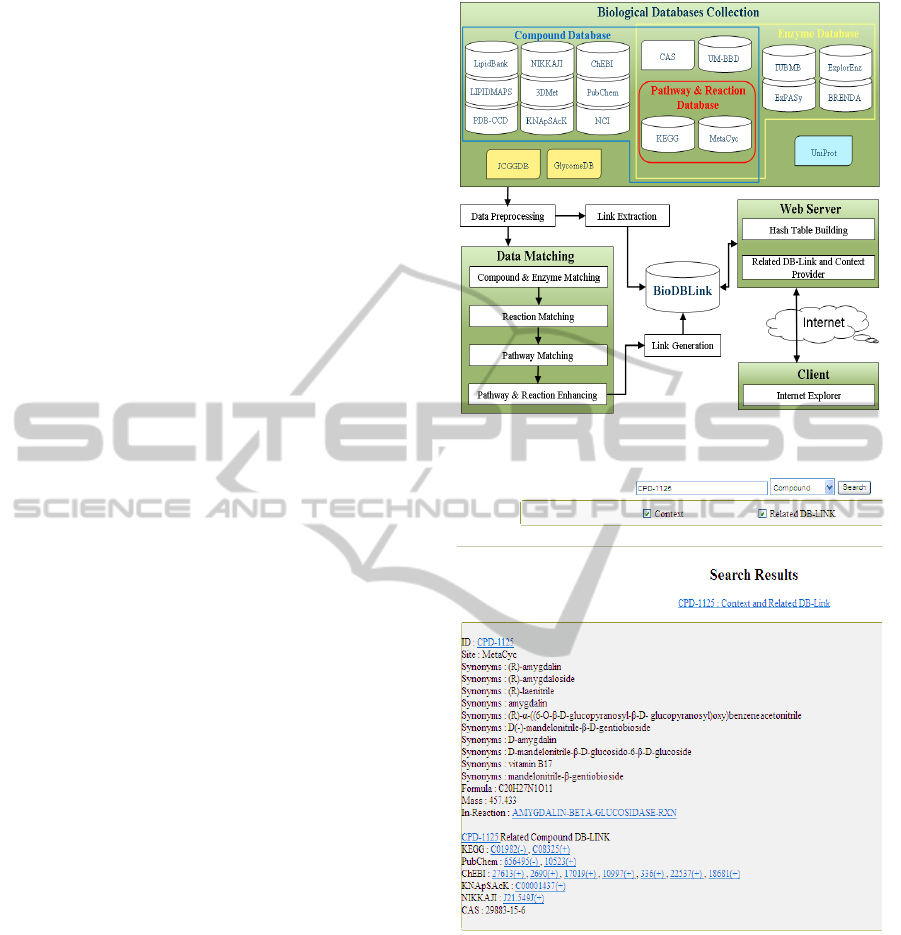
2 BIODBLINK OVERVIEW
As in Figure 1, BioDBLink (Biological Database
Link) is a database link server that automatically
collects and generates cross links among biological
databases. It provides a query interface for searching
compounds, enzymes, reactions and pathways. For
compound query, it accepts a compound name as the
input, and returns a compound description as well as
a list of database links. Figure 2 gives an illustration
of compound query for compound CPD-1125. A
link marked by symbol “(+)” indicates that it is
discovered by data matching but not provided in the
database. Symbol “(-)” indicates that the link is
provided in the database but is suggested to remove
by data matching. For link correction, the old link
will be marked by “(-)”, and a newly discovered link
marked by “(+)” will be added to replace the old one.
The current version of BioDBLink available at
http://140.123.102.75:8080/pathway/index.jsp
provides links generated from the following
databases.
1. Pathway databases: KEGG and MetaCyc.
2. Compound databases: PubChem, ChEBI,
KNApSAcK, LIPIDMAPS, LipidBank, PDB-
CCD, 3DMet, NIKKAJI, NCI, and UM-BBD.
3. Enzyme databases: ExplorEnz, IUBMB,
ExPASy, UM-BBD, and BRENDA.
Figure 1: BioDBLink system overview.
Figure 2: BioDBLink web query interface.
3 BIODBLINK TECHNIQUES
In this section, we present details of our data
matching approach. At first, we download data
descriptions of compounds, enzymes, reactions and
pathways from collected databases. Because those
databases don’t have common data format, we
preprocess those data to a common format.
We then perform a multilevel data matching to
identify data records from different databases that
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
126

describe the same object. We classify information in
pathway databases into 3 levels: compounds,
reactions and pathways. The matching process
consists of two phases: the identification phase and
the enhancing phase. In identification phase, we
identify data matching from lower levels to higher
levels, using matching in lower levels to infer
matching in higher levels. In the enhancing phase,
we use matching identified in higher levels to
enhance matching in lower levels. Finally, we use
our matching result to generate new links and
correct existing links. We next present details of our
data matching methods.
3.1 Compound Matching
We define a formula to evaluate the matching score
between any pair of compound descriptions. Two
compound descriptions are matched if their
matching score is larger than the threshold (1.5 in
our experiment). The compound matching score is a
combination of the name score and formula score.
Given two name sets
1
ns and
2
ns , the name
score
),(
21
nsnsnscore
between them is defined as
the maximum of matching scores between any pair
of names from them.
),(max),(
21
,
21
2211
nnmatchnsnsnscore
nsnnsn ∈∈
=
The matching score between two names is 1.5 if
they are the same string, is 1 if they become the
same after special words, such as -D-, -L-, -O-,
alpha, beta and gamma, are removed, is 0.8 if they
are partially matched, and is 0 otherwise.
To evaluate formula score, we classify and count
the number of atoms in the following 3 types. Type
1 consists of atoms appearing in both formulas with
the same number, type 2 consists of atoms appearing
in both formulas with different numbers, and type 3
consists of atoms appearing in only one of the two
formulas. Consider two formula
1
f
and
2
f . Let
321
,, mmm
denote the number of atoms in type 1, 2
and 3, respectively. The formula score
),(
21
fffscore
is defined as follows.
321
1
21
5.12.1
),(
mmm
m
fffscore
++
=
Given two compound descriptions
),(
111
fnsd =
and
),(
222
fnsd =
, their compound matching score
is defined as follows.
),(),(),(
212121
fffscorensnsnscoreddCompScore +=
3.2 Enzyme Matching
Given two enzyme name sets
1
ens and
2
ens , the
enzyme score
),(
21
ensenseEnzymeScor
between
them is defined as the maximum of matching scores
between any pair of names from them.
),(max),(
21
,
21
2211
enenmatchensenseEnzymeScor
ensenensen ∈∈
=
The matching score between two names is 2.2 if
they are the same string, is 1.2 if they become the
same after special words, such as -D-, -L-, -O-,
alpha, beta and gamma, are removed, and is 0
otherwise.
3.3 Reaction Matching
Each reaction description consists of a set
EC
of
EC-numbers, a set
Cset
of compounds and a set
PCset
of primary compounds. Each compound is
labelled as substrates or products. Let
),,(
1111
PCsetCsetECr =
and
),,(
2222
PCsetCsetECr =
be
two reactions. The reaction matching score
),(
21
rrRScore
is derived from EC-numbers,
compound sets and primary compound sets as
follows.
,),(
),(5.0
),(5.0),(
21
21
2121
CsetCsetCscore
PCsetPCsetPCscore
ECECECscorerrRScore
+×
+
×
=
If
1
PCset
= 0 or
2
PCset
= 0, the reaction matching
score
),(
21
rrRScore
is derives as follows.
,),(5.1
),(5.0),(
21
2121
CsetCsetCscore
ECECECscorerrRScore
×
+×=
),(
21
PCsetPCsetPCscore
, denotes the ratio of
matched compounds in
1
PCset
and
2
PCset
. Let
),(
21
CsetCsetPCM
denote the set of matching pairs
between
1
PCset
and
2
PCset
.
|)||,max(|
|),(|
),(
21
21
21
PCsetPCset
PCsetPCsetPCM
CsetCsetPCscore =
),(
21
CsetCsetCscore
denotes the ratio of matched
compounds in
1
Cset
and
2
Cset
. Let
),(
21
CsetCsetCM
denote the set of matching pairs
between
1
Cset
and
2
Cset .
|)||,max(|
|),(|
),(
21
21
21
CsetCset
CsetCsetCM
CsetCsetCscore =
BIODBLINK: MULTI-LEVEL DATA MATCHING FOR AUTOMATIC GENERATION OF CROSS LINKS AMONG
BIOCHEMICAL PATHWAY DATABASES
127

Each EC-number consists of 4 digital numbers.
The matching score between two EC-numbers is
assigned to 1 if they match all 4 numbers, 0.75 if
they match the first 3 numbers, 0.5 if they match the
first 2 numbers, 0.25 if they match the first number,
and 0 otherwise. The matching score
),(
21
ECECECscore
is defined as the maximum of
matching scores between any pair of EC-numbers in
1
EC
and
2
EC
.
Compounds are labelled as primary substrate,
secondary substrate, primary product or secondary
product. Primary substrates and primary products are
the common substrates and products from one
enzymatic reaction connected to other reactions.
These common substrates are primary substrates,
and the common products are primary products. The
remaining substrates are secondary substrates. The
remaining products are secondary products. In
pathway view, primary compounds are more
important than secondary compounds.
In reaction matching,
Cscore
, the ratio of
matching compounds, is the main measure.
ECscore
and
PCscore
are added to handle missing
data. While compounds are not fully matched due to
compound missing, the score can be enhanced by
matching in EC-numbers or primary compounds.
The matching threshold is 1.5.
Note that, in our data set, there are more than
60% of reactions with primary compounds ratio
larger than 0.5, 60% (5201/8711) in MetaCyc and
61% (4991/8172) in KEGG. This implies that as
long as two reactions have the same set of primary
compounds, their reaction score is often greater than
1.
3.4 Pathway Matching
Each pathway consists of a set of reactions and EC-
numbers. Matching reactions are used to identify
part-of relations between pathways in different
databases as follows. Let
1
D and
2
D be two
pathway databases. Let
p
be a pathway in
1
D . We
want to identify a candidate set
M
of pathways in
2
D for
p
. The candidate set is identified iteratively
as follows. Let
)( pR
be the set of reactions in
p
.
In each iteration, find the pathway
q in
2
D that has
maximum of matching reactions in
)( pR
. Add
q to
M
and remove q from
2
D . Repeat above
process until no more matched reactions exists
between
)( pR
and remaining pathways in
2
D . We
find
Let
)( pEC
and
)(qEC
denote the set of EC-
number in
p
and q , respectively. out pathway
matching from candidate set
M
. The pathway
matching score
),( qpPscore
is derived from the
number of common reactions and EC-numbers as
|))(||,)(min(|
|)()(|
|))(||,)(min(|
|)()(|
qECpEC
qECpEC
qRpR
qRpR ∩
+
∩
Two pathways are matched if their pathway
matching score is larger than the threshold (0.6 in
our experiment).
3.5 Enhancing Compound Matching
and Reaction Matching
For compound matching, the purpose of enhancing
phase is to discover new matching not yet identified
due to different names in different databases, and
remove matching induced errors in the databases.
Reaction matching is used to enhance compound
matching. When two compounds participating in
matched reactions, their compound matching
score,
CompScore
, is increased 0.5. When two
matched compounds do not participate in matched
reaction, their score is decreased.
Pathway matching is used to enhance reaction
matching. As illustrated in Figure 5, supposed in two
matched pathways Pathway1 and Pathway2, R1
matches R1’ and R3 matches R3’. We then increase
the matching score between R2 and R4.
Figure 5:
)4,2( RRRScore
is increased.
4 EXPERIMENT
We experiment our approach over a collection of
popular pathway, compound and enzyme databases,
including KEGG, MetaCyc, PubChem, ChEBI,
KNApSAcK, LIPIDMAPS, LipidBank, PDB-CCD,
3DMet, NIKKAJI, NCI, and UM-BBD, ExplorEnz,
IUBMB, ExPASy, UM-BBD and BRENDA.
The version of MetaCyc which we use is 14.0.
We use APIs provided by KEGG to retrieve
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
128
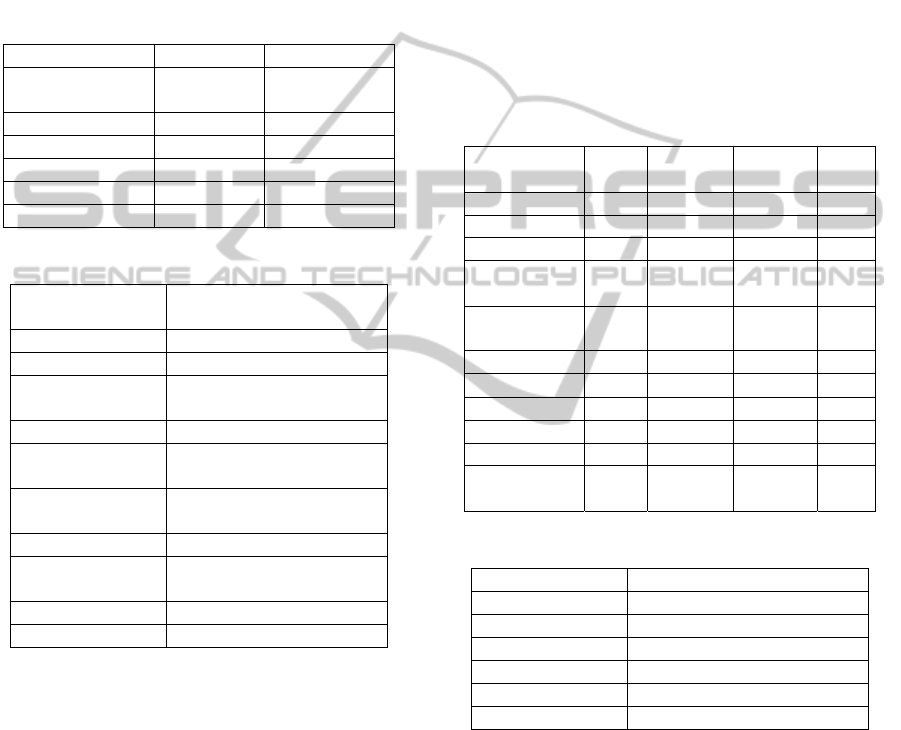
pathway data from KEGG. Table 1 gives statistics of
data collected from MetaCyc and KEGG.
Furthermore, we use compound links collected from
KEGG and MetaCyc to collect data from compound
databases, such as PubChem, ChEBI, KNApSAcK,
LIPIDMAPS, LipidBank, PDB-CCD, 3D-MET,
NIKKAJI, NCI and UM-BBD-CPD. Table 2 gives
statistics of compounds collected from each
compound database.
Table 1: Database statistics between KEGG and MetaCyc.
Database context KEGG MetaCyc
Collected date 2010/06/11 v14.0
(2010/03/18)
Pathway 165 1719
Reaction 8172 8711
Enzyme 5184 9050
Compound 16250 8572
Undefined compound - 1522
Table 2: Compound database statistics.
Compound database Number of collected
compounds (date: 2010/06/13)
PubChem 20711
ChEBI 6574
KNApSAcK 4204 (4246 links provided, but
24 are detected as dead links)
LIPIDMAPS 783
LipidBank 468 (490 links provided, but 22
are detected as dead links)
PDB-CCD 1424 (1449 links provided, but
25 are detected as dead links)
3D-MET 5640
NIKKAJI 6814 (6852 links provided, but
38 are detected as dead links)
NCI 222
UM-BBD-CPD 50
Table 3 gives the number of links predicted by
compound matching, the number of links extracted
from existing databases, and the precision and recall
with respect to extracted links. Our approach
achieves very high recall rate, and is able to identify
more than 90% of cross links provided in current
databases. For compound matching from MetaCyc
to KEGG, our approach identifies 1174 more than
the number of unification links provided by
MetaCyc. This indicates that our approach has the
potential to discover cross links not provided by
current databases.
Table 4 gives statistics of each enzyme database.
For enzyme matching, our experiment identifies
5846 matches from MetaCyc to KEGG. Enzyme
matching result is given in Table 5.
For reaction matching, we identify 4097
matching pairs between MetaCyc and KEGG.
Among them, 264 pairs have different EC-numbers,
and 315 pairs have one reaction missing EC-number.
For those missing EC-numbers, we assign EC-
numbers to them according to their matched
reactions. For those with different EC-numbers, we
suggest them to biologists to check their correct EC-
numbers. Note that MetaCyc v14.0 provides 3260
reaction links and 7 dead links to KEGG. Our
approach identifies 2829 of them, and discovers 7 of
them are invalid. The recall rate is 0.868.
Table 3: Compound matching result.
Databases
predicte
d links
existing
links
Precision
Recall
MetaCyc to KEGG 6129 4244 0.984 0.966
KEGG to PubChem 13652 13607 0.999 0.997
KEGG to ChEBI 6386 5737 0.997 0.961
KEGG to
KNApSAcK
4391 4151 0.992 0.951
KEGG to
LIPIDMAPS
867 782 0.999 0.913
KEGG to LipidBank 699 427 0.982 0.934
KEGG to PDB-CCD 2309 1411 0.984 0.807
KEGG to 3D-MET 5689 5640 0.999 0.994
KEGG to NIKKAJI 7349 6754 0.993 0.974
MetaCyc to NCI 404 222 0.986 0.658
MetaCyc to
UM-BBD-CPD
53 50 1 0.92
Table 4: Enzyme database statistics.
Enzyme database Number of collected enzymes
Collected Date 2010/06/13
ExplorEnz 4257
IUBMB 4257
ExPASy 4257
UM-BBD 289
BRENDA 4257
For pathway matching, each matching denotes a
part-of relation. Our approach identifies 1343
pathway relations. Among them, 1218 are one-to-
one and 125 are one-to-many. MetaCyc v14.0
provided 24 pathway unification links to KEGG.
Our approach identifies 15 of them, and discovers 4
of them are invalid. The recall rate is 0.75. If we set
pathway matching threshold to 0.2, we can identify
1484 pathway matchings. Among them, 851 are one-
to-one and 633 are one-to-many. We identify 16
links that MetaCyc provided, recall rate is 0.8.
BIODBLINK: MULTI-LEVEL DATA MATCHING FOR AUTOMATIC GENERATION OF CROSS LINKS AMONG
BIOCHEMICAL PATHWAY DATABASES
129

Table 5: Enzyme matching result.
From DB 1 to DB 2
Number of
predicted
links
Number of
extracted links
Recall w.r.t.
extracted links
MetaCyc to KEGG 5855 7597 0.74
KEGG to ExplorEnz 4254 4257 0.999
KEGG to IUBMB
4253 4257 0.999
KEGG to ExPASy
4204 4257 0.99
KEGG to UM-BBD 318 289 0.98
KEGG to BRENDA 4155 4257 0.97
5 CONCLUSIONS
In this paper, we present a pathway database link
server BioDBLink that can automatically collect and
generate cross links among biological databases. The
core of BioDBLink is a multi-level data matching
technique that identifies and matches data records or
elements describing the same object. Matching
results can also be used to induce more accurate and
complete object descriptions, remove data
redundancy, and check data consistency. Experiment
on a set of pathway, compound and enzyme
databases shows that our approach is feasible,
identifies a large number of matchings, and detect
database inconsistency and duplicate errors. In the
future, we will continue to extend our server to
incorporate more databases available on internet,
and develop data matching techniques to match
other types of biological entities. Our goal is to
provide a database link server for more biological
databases.
REFERENCES
Birkland A., Yona G., 2006. BIOZON: a system for
unification, management and analysis of
heterogeneous biological data. In BMC Bioinformatics.
7:70doi:10.1186/1471-2105-7-70.
Garcia C. A., Chen Y. P., Ragan M. A.,2005. Information
integration in molecular bioscience. In Applied
Bioinformatics, 4(3), 157-173.
Macauley J., Wang H., Goodman N., 1998. A Model
System for Studying the Integration of Molecular
Biology Databases. Bioinformatic, 14(7), 575-582.
Krishnamurthy L., Nadeau J., Ozsoyoglu G., Ozsoyoglu
M., Schaeffer G., Tasan M. and Xu W., 2003.
Pathways database system: an integrated system for
biological pathways. Bioinformatic, 19(8), 930-937.
Chen Y. P., Chen Q., 2006. Analyzing Inconsistency
Toward Enhancing Integration of Biological
Molecular Databases. In APBC , thefourth Asia-
Pacific Bioinformatics Conference, 197-206.
Rajasimha K. H., 2004. PathMeld: A Methodology for the
Unification of Metabolic Pathway Databases.
Computer Science and Application, 2004
Jyh-Jong Tsay, Bo-Liang Wu and Chien-Wen Chen. Data
Matching for Physical Integration of Biochemical
Pathway Databases. IEEE International Conference
on Bioinformatics and Bioengineering, 2009.
Lim E., Chiang R. H., 2000. The integration of
relationship instances from heterogeneous databases.
Decision Support Systems, 29, 153-167
Sujansky W.,2001. Heterogeneous Database Integration in
Biomedicine. Journal of Biomedical Informatics, 34,
285-298.
Li W. and Clifton C., 2000. SEMINT: A tool for
identifying attribute correspondences in heterogeneous
databases using neural networks. Data and Knowledge
Engineering, 33, 49-84.
KEGG, Available at http://www.genome.jp/kegg/
Karp P. D., Riley M., Saier M., Paulsen I., Paley S., and
A, 2000. Pellegrini-Toole, The EcoCyc and MetaCyc
databases. Nucleic Acids Research, 28, 56-59.
METACYC, Available at http://metacyc.org/
Green M. L. and Karp P. D., 2005. Genome annotation
errors in pathway databases due to semantic ambiguity
in partial EC numbers. Nucleic Acids Research,
33(13), 4035-4039.
PubChem, Available at http://pubchem.ncbi.nlm.nih.gov/
ChEBI, Available at http://www.ebi.ac.uk/
KNApSAcK, Available at http://kanaya.aist-nara.ac.jp/
LIPIDMAPS, Available at http://www.lipidmaps.org/
LipidBank, Available at http://lipidbank.jp/
PDB-CCD, Available at http://remediation.wwpdb.org/
3DMET, Available at http://www.3dmet.dna.affrc.go.jp/
Nikkaji, Available at http://nikkajiweb.jst.go.jp/
NCI, Available at http://cactus.nci.nih.gov/
UM-BBD, Available at http://umbbd.msi.umn.edu/
ExplorEnz, Available at http://www.enzyme-database.org/
IUBMB, http://www.chem.qmul.ac.uk/iubmb/enzyme/
ExPASy, Available at http://www.expasy.org/
BRENDA, Available at http://www.brenda-enzymes.org/
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
130
