
A COMPUTATIONAL STRATEGY TO INVESTIGATE
RELEVANT SIMILARITIES BETWEEN VIRUS
AND HUMAN PROTEINS
Local High Similarities between Herpes and Human Proteins
Anna Marabotti
Istituto di Tecnologie Biomediche, CNR, Segrate (MI), Italy
Istituto di Scienze dell’Alimentazione, CNR, Avellino, Italy
Corrado Cirielli, Daniela Agnese D’Arcangelo
Istituto Dermopatico dell’Immacolata, IDI-IRCCS, Rome, Italy
Claudia Giampietri
Department of Histology and Medical Embryology, “Sapienza” University of Rome, Italy
Francesco Facchiano
Dipartimento Ematologia, Oncologia e Medicina Molecolare, Istituto Superiore di Sanità, Rome, Italy
Antonio Facchiano
Istituto Dermopatico dell’Immacolata, IDI-IRCCS, Rome, Italy
Angelo M. Facchiano
Istituto di Scienze dell’Alimentazione, CNR, Avellino, Italy
Keywords: Proteome comparison, Molecular mimicry, Autoimmunity, Local similarity.
Abstract: Investigating primary sequence and structural features of viral proteins/genes has revealed molecular
mimicry and evolutionary relationship linking viruses to eukaryotes. The continuous improvement in
sequencing-techniques makes available almost daily the whole genome/proteome of several
microorganisms, making now possible systematic analyses of evolutionary correlations and accurate
phylogeny investigations. In the present study we set up a methodology to identify significant and relevant
similarities between viral and human proteomes. To this aim, the following steps were applied: i)
identification of local similarity corresponding to continuous identity over at least 8-residues long
fragments; ii) filtering results for statistical significance of the identified similarities, according to BLAST
parameters for short sequences; iii) additional filters applied to the BLAST outputs, to select specific
viruses. The present study indicates a novel accurate methodology to find relevant similarities among virus
and human proteomes, useful to further investigate pathogenic mechanisms underlying infectious and non-
infectious diseases.
1 INTRODUCTION
Several studies investigate genetic predisposition
toward human disorders (Baranzini, 2009; Rubstov,
2010). Mechanisms involving a genetic basis may
require events occurring at the germinal level
(giving hereditary disorders) or at somatic levels.
Environmental stimuli and life style, such as smoke
183
Marabotti A., Cirielli C., Agnese D’Arcangelo D., Giampietri C., Facchiano F., Facchiano A. and M. Facchiano A..
A COMPUTATIONAL STRATEGY TO INVESTIGATE RELEVANT SIMILARITIES BETWEEN VIRUS AND HUMAN PROTEINS - Local High Similarities
between Herpes and Human Proteins.
DOI: 10.5220/0003156801830188
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2011), pages 183-188
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

habit, professional chemical exposure, diet style or
exposure to pollution or to infectious agents, may
interfere at the genetic level on the system
homeostasis within the human body. In several cases
infective agents, besides the direct infection disease,
may trigger additional mechanisms responsible of
the onset of additional diseases. Virus infections are
often responsible of additional serious pathologies,
and in most cases the mechanisms underlying the
onset of these “secondary” pathologies are not fully
elucidated. For instance, Human Papilloma virus
(HPV) infection is known to strongly relate to the
occurrence of cervix cancer in humans (Leggatt,
2007); helicobacter pylori infection is strongly
related to gastric ulcer and gastric cancer (Cao,
2007); hepatitis B virus (HBV) infection has been
proposed to have a role in the development of
hepatocellular carcinoma (Neuveut, 2010); diabetes
has been related with rotavirus infection (Maklea,
2006) or coxsackie virus infection (Peng, 2006);
bacterial infections have been shown to be related to
heart diseases (Ott, 2006); myocarditis has been
proposed to have an autoimmune basis related to
auto-antibodies against cardiac myosin and based on
a mimicry process between cardiac myosin and the
beta-adrenergic receptor (Li, 2006); Herpes Virus
type 7 has been shown to be involved in initiation
and maintenance of Graves’ disease autoimmune
process (Leite, 2010); Herpes Virus type 1 infection
has been strongly related to autoimmune reaction
based on the auto-reactive T lymphocytes
recognizing shared epitopes (Zhao, 1998); herpes
antigens have been suggested to play a molecular
mimicry role for autoimmune-based diseases such as
psoriasis (Kirby, 2000; Mehraein, 2004) and
antibodies against Herpes viruses have been found in
immuno-deficient or auto-immune patients (Thomas,
2008). A recent study (Fumagalli, 2010) pointed out
that up to 8% of the human genome is of viral
source, representing the “fossil remnants of past
infections” via integration at several sites. Consistent
with these findings, a strong rational supports the
investigation of human-to-viral structural-functional
relationships based, at least in some cases, onto a
common-antigen sharing process. We have
previously suggested unexpected evolutionary
relations between human-lymphocytes CD4 receptor
and his counterpart HIV-capsid-constituent GP120
(Facchiano, 1995; Facchiano, 1996); allele
variability has been related to viral infection and
susceptibility, with a significant association of HIV
progression with patients HLA status (Limou, 2009;
Fellay, 2007). Based on significant proteins
similarities we have also suggested the occurrence of
molecular mimicry between pathogens and human
proteins possibly underlying different diseases
(Benvenga, 1995; Benvenga, 1999; Benvenga,
2003). According to such large body of evidence, it
is now accepted that eukaryotes have been, and still
are, under a strong selective pressure, driven, at least
in part, by viral/prokaryotes infections, with human
gene variants associated to pathogens–related
infections. The strong improvement of the
sequencing techniques is producing a continuous
novel identification of the entire genome/proteome
of several micro-organisms, including viral
pathogens. Such sequences, or at least those
available freely on the net, can be analyzed with a
number of different approaches and software to
investigate structural features as well as local or
general evolutionary correlations. In the current
study we present a novel accurate methodology
which allows to identify relevant high local
similarities between viral entire proteome and
human proteome.
2 METHODS
Protein sequences from viral and human source were
obtained by the RefSeq database to constitute our
viral and human proteome databases, which included
82918 and 39037 protein sequences, respectively.
The two databases were initially compared by using
self-developed proprietary PERL scripts. The
comparison was performed in two steps: in the first
step all 8 amino acids-long fragments were extracted
from the viral database; each virus fragment was
then searched in the human database. Any time a full
identity was found (8 identities out of 8), extensions
at the N- or C- terminus were investigated to verify
the occurrence of identity over a longer fragment.
The second step of the analysis was then carried
out with BLASTplus (available at the NCBI web
site: http://www.ncbi.nlm.nih.gov/) to verify the
statistical significance of the identities found. All
viral fragments selected within the first step were
compared to the human proteome database in order
to obtain scores and significance evaluation. The
“blastp-short” settings were applied, i.e. parameters
were optimized for query sequences shorter than 30
residues. Namely, the following settings were
applied: word_size=2, gapopen=9, gapextend=1,
matrix=PAM30, threshold=16, comp_based_stats=2.
Finally, a final filter based on the virus name was
applied to extract the similarities identified by
BLAST, concerning protein sequences from specific
viruses.
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
184

3 RESULTS AND DISCUSSION
During the first step of our search strategy, we
extracted all fragments of 8 amino acids from the
viral proteins. This minimum length was chosen as a
threshold to select fragments with a putative activity
as antigenic peptides, as discussed below. These
fragments were searched in the human proteome
database, and extended when possible to the
maximum length of identical segment. The search
identified a very high number of non redundant
fragments, i.e. 42993 having identical sequence in
viral and human proteins, with length of 8 or more
amino acids. In many cases, the sequences are
characterized by low sequence complexity, i.e.,
composed by only one or two different residues. The
statistical relevance of the similarity involving very
short sequences or low-complexity fragments is
often considered very low or of difficult evaluation.
However, highly repetitive sequences are known to
play several key functions: for example, collagen
and sericin (the protein composing the silk) are
characterized by low-complexity sequences; further,
proline-rich peptides have low-complexity but
represent key sequences recognized by SH3 domain
(Yu, 1994); a 9 residues long poly-arginine has been
demonstrated to play a key role in cellular uptake
(Wender, 2000); proline-enrichment within peptide-
sequence may increase binding to mitochondrial
targets (Serasinghe, 2010); leucine-rich or lysine-
rich fragments have shown adhesive-, heparin-
binding and DNA-binding features. In order to
follow a statistics-driven approach, we decided to
proceed the analysis using only BLAST-defined
statistically significant identities; therefore, in the
second step of the present procedure we compared
any selected virus-fragment to the human proteome
by BLASTplus, using statistical significance settings
specifically chosen for short sequences. This second
step selected 1076 viral fragments which share
statistically significant similarity with human protein
sequences and constituted the VISHUM database
(VIral Similarities to HUMan fragments). This is
one of the products of the present study; it is a
growing database, planned to be updated
periodically for the presence of new available viral
sequences, as well as extended to sequences
extracted from different databases such as GenBank
and Uniprot. It presently contains the sequences and
the cross-references to the original viral and human
databases. Further investigation is now been carried
out on the VISHUM database; as an example, we
selected from the VISHUM database the herpesvirus
fragments and sorted the results by sequence
identity. The herpesvirus selected fragments
showing at least 8 continuous identities and at least
90% of sequence identity with human proteins are
listed in Table 1. In all cases, with just one
exception, the extension of the peptide is longer or
much longer than the minimum requested threshold.
While a simpler search strategy could be applied,
nevertheless we aimed at finding results significant
from both structural and functional point of view, to
select fragments with a full identity for a minimum
length of 8 amino acids (as minimum requirement
for a putative antigenic activity), further extended to
the maximum length of identity or similarity; the
BLAST evaluation of scores and significance was
then applied to each fragment to fix the best level of
extension.
To our knowledge this study reports for the first
time a systematic approach to investigate structural-
functional correlations of viral proteins with human
proteins. Choosing 8 consecutive residues, as the
minimum requested identity, represented the way to
wipe out all short identities difficult to evaluate from
the statistical and functional point of view, and
allowed to select those with high statistical
significance and with an immune-related biological
role. In fact, antigen presenting cells (APCs) carry 8-
residues long peptides to lymphocytes to elicit the
immune response. Additional post-filtering may
further improve the fragment selection procedure
and the further ongoing investigation of the
VISHUM database. For instance only human-
specific viral fragments may be selected to be
compared to human sequences or both human-
specific and human-non-specific fragments can be
selected to implement evolutionary-based analyses.
Moreover, our study can also be compared to data
collected from public databases related to human –
virus interactions. We present here data obtained by
a post-filtering procedure based on virus name (see
Table 1); other filters may be implemented within
this strategy, such as, for an example, a 3D-filter to
select fragments with a known 3D structure.
The procedure presented in the current study
allows to identify viral fragments sharing full
identity with human proteins, with a strong
statistical significance. The occurrence of highly
significant local identities may indicate specific
regions with common ancestry or regions where a
local evolutionary pressure or a molecular mimicry
process may have occurred. A very recent report
underlines the role of viral infections as possible
trigger of autoimmune disorders in genetically
predisposed individuals (Ji, 2010); hence developing
VISHUM database and effective procedures to
A COMPUTATIONAL STRATEGY TO INVESTIGATE RELEVANT SIMILARITIES BETWEEN VIRUS AND
HUMAN PROTEINS - Local High Similarities between Herpes and Human Proteins
185
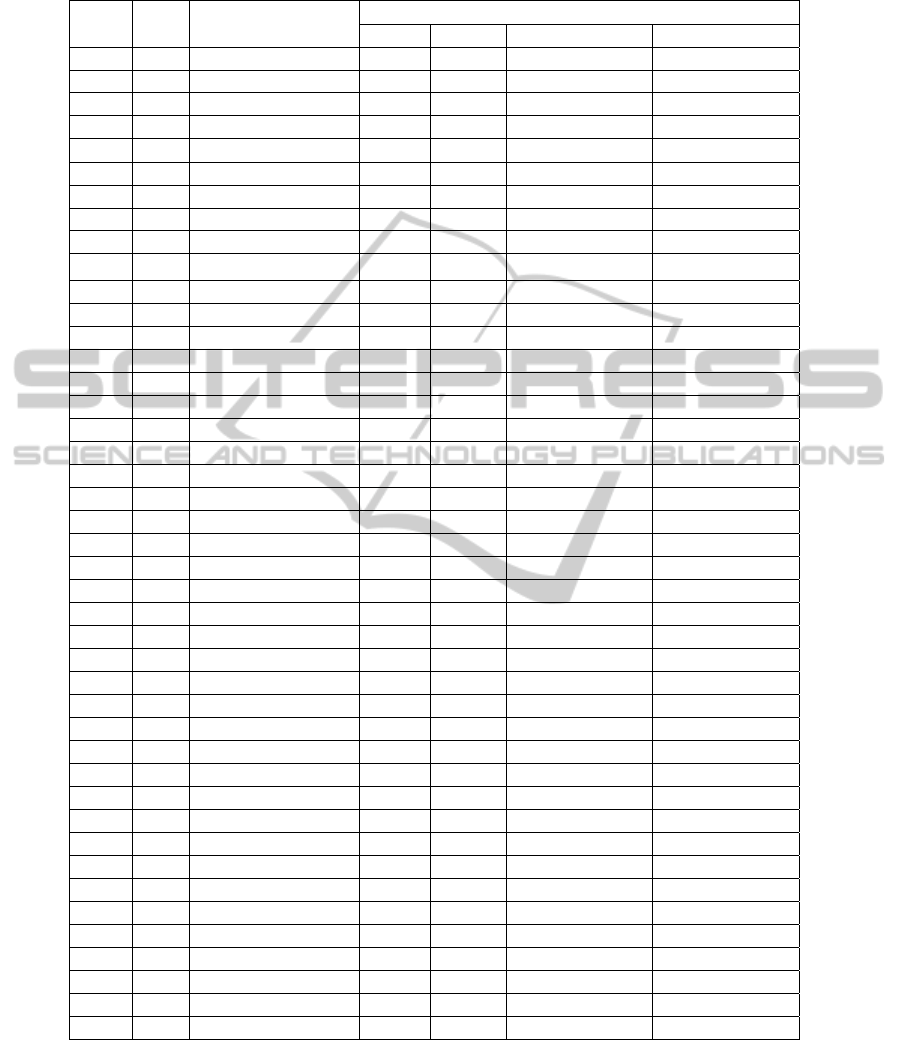
Table 1: Results generated by the search strategy, filtered for human herpesviruses. The best hits are shown, in terms of
percentage of identity (at least 90%).
Peptide
Code
Length Virus name
BLAST results
Score E-value Number of matches Percentage of identity
10932 11 Human herpesvirus 5 26.2 3.8 11 100
18349 42 Human herpesvirus 4 type 2 87.8 2.00E-18 42 100
18349 42 Human herpesvirus 4 87.8 2.00E-18 42 100
22175 13 Human herpesvirus 4 type 2 30.4 0.19 12 100
22175 13 Human herpesvirus 4 30.4 0.19 12 100
22175 13 Human herpesvirus 8 28.5 0.8 11 100
28804 10 Human herpesvirus 8 25 6.4 10 100
34315 11 Human herpesvirus 8 26.6 2.5 11 100
34752 14 Human herpesvirus 6 35.8 0.006 14 100
34752 14 Human herpesvirus 6 35.8 0.006 14 100
34752 14 Human herpesvirus 7 35 0.01 14 100
35947 12 Human herpesvirus 8 28.5 0.66 11 100
4113 12 Human herpesvirus 8 28.5 0.72 12 100
41176 20 Human herpesvirus 8 44.7 1.00E-05 20 100
43174 14 Human herpesvirus 8 32.3 0.055 14 100
44922 9 Human herpesvirus 2 24.6 7.3 9 100
46915 11 Human herpesvirus 8 31.6 0.089 11 100
53339 31 Human herpesvirus 4 type 2 63.9 3.00E-11 31 100
53339 31 Human herpesvirus 4 63.9 3.00E-11 31 100
5707 9 Human herpesvirus 5 24.3 10 9 100
57304 13 Human herpesvirus 6 28.9 0.55 12 100
6005 8 Human herpesvirus 2 24.3 8.3 8 100
62235 24 Human herpesvirus 8 55.8 9.00E-09 24 100
55656 39 Human herpesvirus 4 type 2 45.8 9.00E-06 38 97
55656 39 Human herpesvirus 4 45.8 9.00E-06 38 97
58282 20 Human herpesvirus 8 43.5 4.00E-05 18 94
40584 15 Human herpesvirus 3 34.7 0.014 14 93
40584 15 Human herpesvirus 8 33.1 0.041 14 93
34752 14 Human herpesvirus 5 34.7 0.013 13 92
35527 15 Human herpesvirus 8 33.5 0.028 13 92
41386 14 Human herpesvirus 4 33.5 0.026 13 92
41386 14 Human herpesvirus 4 type 2 33.5 0.026 13 92
57304 13 Human herpesvirus 8 32.3 0.05 12 92
57304 13 Human herpesvirus 4 type 2 32.3 0.05 12 92
57304 13 Human herpesvirus 4 32.3 0.05 12 92
35158 23 Human herpesvirus 4 type 2 45.4 1.00E-05 21 91
35158 23 Human herpesvirus 4 45.4 1.00E-05 21 91
57304 13 Human herpesvirus 6 27.3 1.8 11 91
62235 24 Human herpesvirus 3 53.5 4.00E-08 22 91
16190 22 Human herpesvirus 8 48.5 1.00E-06 20 90
40505 21 Human herpesvirus 8 45.4 1.00E-05 19 90
41737 10 Human herpesvirus 4 type 2 26.6 2.1 9 90
41737 10 Human herpesvirus 4 26.6 2.1 9 90
investigate local protein regions may represent a step
forward the improved understating of mechanisms
underlying virus-related disorders.
4 CONCLUSIONS
The present study reports a novel procedure to
identify relevant similarities among virus and human
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
186

proteomes, useful to investigate structural/functional
features underlying infectious and non-infectious
diseases.
ACKNOWLEDGEMENTS
The present project was partially supported by
Programma Italia-USA “Oncoproteomica” (Ref.
527B/2A/5), Programma Italia-USA
“Farmacogenomica Oncologica” (Ref. 527/A/3A/5)
and by Biophysikalische Informations-Therapie
(Freiburg). We also thank Ruth Beisler for the
enthusiastic, stimulating and helpful support to the
entire project.
REFERENCES
Baranzini, S. E., The genetics of autoimmune diseases: a
networked perspective. Curr. Opin. Immunol. 2009.
21: 596-605.
Rubtsov, A. V., Rubtsova, K., Kappler, J. W., and P.
Marrack. Genetic and hormonal factors in female-
biased autoimmunity. Autoimmun. Rev. 2010. 9:494-8
Leggatt, G. R. and I. H Frazer. HPV vaccines: the
beginning of the end for cervical cancer. Curr. Opin.
Immunol. 2007. 19: 232-238.
Cao, X., Tsukamoto, T., Nozaki, K., Tanaka, H., Cao, L.,
Toyoda, T. et al.. 2007 Severity of gastritis determines
glandular stomach carcinogenesis in Helicobacter
pylori-infected Mongolian gerbils. Cancer Sci. 98:
478-483.
Neuveut, C., Wei, Y., and M. A. Buendia. Mechanisms of
HBV-related hepatocarcinogenesis. J. Hepatol. 2010.
52:594-604.
Makela, M., Vaarala, O., Hermann, R., Salminen, K.,
Vahlberg, T., Veijola, R. et al. 2006 Enteral virus
infections in early childhood and an enhanced type 1
diabetes-associated antibody response to dietary
insulin. J. Autoimmun. 27:54-61.
Peng, H. and W. Hagopian. 2006 Environmental factors in
the development of Type 1 diabetes. Rev. Endocr.
Metab. Disord. 7:149-162.
Ott, S. J., El Mokhtari, N. E., Musfeldt, M., Hellmig, S.,
Freitag, S., Rehman, A. et al. 2006 Detection of
diverse bacterial signatures in atherosclerotic lesions
of patients with coronary heart disease. Circulation
113:929-937.
Li, Y., Heuser, J. S., Cunningham, L. C., Kosanke, S. D.,
and M. W. Cunningham. 2006 Mimicry and Antibody-
Mediated Cell Signaling in Autoimmune Myocarditis.
J. Immunol. 177:8234-8240.
Leite, J. L., Bufalo, N. E., Santos, R. B., Romaldini, J. H.,
and L. S. Ward. 2010 Herpesvirus type 7 infection
may play an important role in individuals with a
genetic profile of susceptibility to Graves' disease.
Eur. J. Endocrinol. 162:315-321.
Zhao, Z. S., Granucci, F., Yeh, L., Schaffer, P. A., and H.
Cantor. 1998. Molecular Mimicry by Herpes Simplex
Virus-Type 1: Autoimmune Disease After Viral
Infection. Science 279:1344-1347.
Kirby, B., Al-Jiffri, O., Cooper, R. J., Corbitt, G., Klapper,
P. E., and C. E. Griffiths. 2000. Investigation of
cytomegalovirus and human herpes viruses 6 and 7 as
possible causative antigens in psoriasis. Acta Derm.
Venereol. 80:404-406.
Mehraein, Y., Lennerz, C., Ehlhardt, S., Zang, K. D. and
H. Madry. 2004. Replicative multivirus infection with
cytomegalovirus, herpes simplex virus 1, and
parvovirus B19, and latent Epstein-Barr virus infection
in the synovial tissue of a psoriatic arthritis patient. J.
Clin. Virol. 31:25-31.
Thomas, D., Karachaliou, F., Kallergi, K.,
Vlachopapadopoulou, E., Antonaki, G., Chatzimarkou,
F. et al. 2008. Herpes virus antibodies seroprevalence
in children with autoimmune thyroid disease.
Endocrine 33:171-175.
Fumagalli, M., Pozzoli, U., Cagliani, R., Comi, G. P.,
Bresolin, N., Clerici, M., and M. Sironi. 2010.
Genome-Wide Identification of Susceptibility Alleles
for Viral Infections through a Population Genetics
Approach. PLoS Genet. 6:e1000849.
Facchiano A. 1995. Investigating Hypothetical Products of
Non-coding Frames (HyPNoFs), J. Mol. Evol. 40:570-
577.
Facchiano A. 1996. Coding in noncoding frames. Trends
in Genetics 12:168-169.
Limou, S., Le Clerc, S., Coulonges, C., Carpentier, W.,
Dina C., Delaneau, O., et al. 2009. Genomewide
association study of an AIDS-nonprogression cohort
emphasizes the role played by HLA genes (ANRS
Genomewide Association Study 02). J. Infect. Dis.
199:419-426.
Fellay, J., Shianna, K. V., Ge D., Colombo, S.,
Ledergerber, B., Weale, M. et al. 2007. A whole-
genome association study of major determinants for
host control of HIV-1. Science 317:944-947.
Benvenga, S., and A. Facchiano, 1995. Homology of atrial
natriuretic protein with the proteins associated with
amyloidosis. J. Int. Medicine 237:525-526.
Benvenga, S., Alesci, S., Trimarchi, F., and A. Facchiano.
Homologies of the thyroid sodium-iodide symporter
with bacterial and viral proteins. Endocrinol. Invest.
1999. 22: 535-540.
Benvenga, S., Trimarchi, F., and A. Facchiano. 2003.
Cogan’s syndrome as an autoimmune disease. The
Lancet 361:530-53l.
Yu, H., Chen, J. K., Feng, S., Dalgarno, D. C., Brauer, A.
W., and S. L. Schreiber. 1994. Structural basis for the
binding of proline-rich peptides to SH3 domains. Cell
76:933-945.
Wender, P. A., Mitchell, D. J., Pattabiraman, K., Pelkey,
E. T., Steinman, L., and J. B. Rothbard. The design,
synthesis, and evaluation of molecules that enable or
enhance cellular uptake: peptoid molecular
A COMPUTATIONAL STRATEGY TO INVESTIGATE RELEVANT SIMILARITIES BETWEEN VIRUS AND
HUMAN PROTEINS - Local High Similarities between Herpes and Human Proteins
187

transporters. Proc. Natl. Acad. Sci. U S A. 2000. 97:
13003-13008.
Serasinghe, M. N., Seneviratne, A. M., Smrcka, A. V., and
Y. Yoon. 2010. Identification and characterization of
unique proline-rich peptides binding to the
mitochondrial fission protein hFis1. J. Biol. Chem.
285:620-630.
Ji, Q., Perchellet A., and Goverman J. M. 2010 Viral
infection triggers central nervous system
autoimmunity via activation of CD8+ T cells
expressing dual TCRs. Nature Immunology 11: 628-
635
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
188
