
GEOMETRICAL CONSTRAINTS FOR LIGAND POSITIONING
Virginio Cantoni, Alessandro Gaggia, Riccardo Gatti and Luca Lombardi
University of Pavia, dept. of Computer Engineering and Systems Science,Via Ferrata 1, Pavia, Italy
Keywords: Protein-ligand interaction, Active sites detection, Extended Gaussian Image, Alignment of biological
molecules, Structural matching, Mathematical morphology.
Abstract: The purpose of the activity here described is the morphological and subsequently the geometrical and
topological analysis of the active sites in protein surfaces for protein-ligand docking. The approach follows
a sequence of three steps: i) the solvent-excluded-surface is analyzed and segmented in a number of pockets
and tunnels; ii) the candidate binding sites are detected through a structural matching of pockets and ligand,
both represented through a suitable Extended Gaussian Image modality; iii) the loci of compatible positions
of the ligand is identified through mathematical morphology. This representation of ligand and candidate
binding pockets, the comparison of the morphological similarity and the identification of potential ligand
docking are the novelties of this proposal.
1 INTRODUCTION
Much work has been done on the identification, the
localization, and the analysis of the binding sites of
proteins. The aim, for docking applications, is the
search of sub-regions that are complementary (that is
with concave and convex segments that match each
others) between different molecules. When we have
a large molecule (receptor) and a small molecule
(ligand), docking takes place in a protein cavity;
instead the protein-protein case is usually different,
in fact the docking site is, in general, more planar
than a cavity and the interface has different
characteristics.
In this connection the first sub-problem to be solved,
in protein-ligand interfaces, is to develop the
representations and the data structures suitable to
support the computational methods which consent a
quantitative evaluation of the protein-protein and in
particular the protein-ligand matching on the basis
mainly of their 3D structure. Until now this problem
has been pursued by 'ad hoc' descriptors of patterns
like spin image (Shulman-Peleg, 2004), (Bock,
2007), context shape (Frome, 2004) and harmonic
shape (Glaser, 2006). Some of these approaches are
point-based and in general they look to us
cumbersome with difficulties for management and
processing.
In this paper a new method for representing the
molecule is proposed, and the correspondent data
structure based on a first order statistic of the
orientation is introduced. After the segmentation of
the protein solvent excluded surface (SES) (Cantoni,
2010), the interface regions, which potentially can
be active sites, are represented by a kind of
Extended Gaussian Image (EGI) (Horn, 1984). The
EGI represents the histogram of the orientations
placed on the unitarian sphere and constitutes a
compact and effective representation of a 3D object
as a protein or one of its parts.
This paper is organized as follows: section two
shows a survey of the EGI representations; in
section three, the construction of CE-EGI is
introduced; in section four is introduced the practical
implementation and the data structure; in section
five is described the matching problem and the
possible discriminant functions based on different
distance definition; in section six the detection of
candidate positions in discussed; and finally in
section seven the results for the new solution
proposed for the identification of binding sites,
together with a practical case are presented. The
final section, provides a few concluding remarks and
briefly describes our planned activity in the near
future.
2 RELATED WORKS
The EGI was introduced for applications of photo
204
Cantoni V., Gaggia A., Gatti R. and Lombardi L..
GEOMETRICAL CONSTRAINTS FOR LIGAND POSITIONING.
DOI: 10.5220/0003166002040209
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2011), pages 204-209
ISBN: 978-989-8425-36-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

-metry by B.K.P. Horn (Horn, 1984) in the years '80
and has been extended by K. Ikeuci (the Complex-
EGI) (Kang, 1993) in the years '90 to overcome the
ambiguity rised in the representations by the convex
parts. Later other improvements have been
introduced in sequence: the More Extended
Gaussian Image (MEGI) in 1994, the Multi-Shell
Extended Gaussian Image (MSEGI) and the
Adaptive Volumetric Extended Gaussian Image (A-
VEGI) in 2007, and finally the Enriched Complex
Extended Gaussian Image (EC-EGI) in 2010.
They have not up-to-now been applied on
proteomics; starting from these, we propose a
representation suitable for describing the matching
between the ligand (here the protuberance) and the
protein (the cavity under analysis).
Extended Gaussian Image (EGI). The EGI of a
3D object or shape is an orientation histogram that
records the distribution of surface area with respect
to surface orientation. Each surface patch is mapped
to a point on the unit Gaussian sphere according to
its surface normal. The weight for each surface
normal (represented by a point on the Gaussian
sphere) is the total sum of area of all the surface
patches that are of that surface normal. Being a
distribution related to surface orientation, EGI is in
principle invariant to translation.
Complex EGI (Kang, 1991). CEGI encodes each
surface patch’s signed perpendicular distance from
the reference coordinate center.
It uses a complex number, as opposed to a scalar
in EGI, as the weight for the corresponding point on
the Gaussian sphere. The magnitude and phase of
the complex number are the area and signed
perpendicular distance of the patch (from the origin
of the reference coordinate frame), respectively. The
use of complex numbers allows the area and position
information to be decoupled. Furthermore, the
translation component of the pose can be determined
more readily.
More Extended Gaussian Image (MEGI)
(Matsuo, 1994). The MEGI model consists of a set
of position vectors X
i
for surfaces originating from
an object center and their normal vectors p
i
. Each
length of a normal vector also corresponds with
surface area, as in the EGI. Also this model is shift-
invariant since it is expressed by an object-oriented
coordinate. The MEGI model is an extended EGI
modeling which is able to represent concave objects.
Multi-Shell Extended Gaussian Image (MSEGI)
(Wang, 2007) or Volumetric Extended Gaussian
Image (VEGI) (Zhang, 2006). The VEGI captures
the volumetric distribution of a triangulated 3D
model by connecting the vertices of each triangle
with the geometry centroid of the object to form a
tetrahedron as the elementary volume unit. Then the
3D model is decomposed into a number of N
s
concentric spheres. Each sphere surface is
subdivided in cells, each one identified through their
polar and longitudinal angles (θ
i
, ϕ
j
). The quantized
volume of each tetrahedron and its associated
direction (the outward surface normal) are mapped
to the corresponding cell of the concentric sphere
with radius ρ, obtaining N
s
spherical distribution
functions η (ρ, θ, ϕ). These functions are expanded
into spherical harmonics to achieve a features
vector. The VEGI and this representation, without
canonical alignment, maintains the property of
translation, scaling, rotation invariance and facilitate
multiple scale approximation. An improvement to
fix the irregular sampling of the polar and
longitudinal coordinate system (in the poles there is
a higher sampling density than in the equator) has
been proposed with the Adaptive Volumetric
Extended Gaussian Image (A-VEGI) (Wang, 2007).
Enriched CEGI (Hu, 2010). The EC-EGI
encodes each surface patch’s signed with its 3D
position. It uses three complex numbers, as the
weight for the corresponding point on the Gaussian
sphere. The resultant weight at the point is then the
sum of the contributions of all surface patches that
are of the corresponding surface normal referred to
each one of the coordinate planes. The magnitude
part of the EC-EGI representation is translation-
invariant. This is an important property that allows
the rotation part of pose, in the pose estimation
application, to be determined separately from the
translation. The EC-EGI can be viewed as three
independent Gaussian spheres, each encoding the 3D
position information along the x-, y- and z-axes,
respectively.
In this paper we propose the adoption of this last
EC-EGI for ligand and cavity to evaluate
quantitatively the matching between candidate active
sites and ligand. ρ
3 CONSTRUCTION OF CE-EGI
A given 3D molecule, modeled through its Solvent
Excluded Surface in a triangular mesh, is described
by the set of triangles:
=
{
,…,
}
,
⊂
(1)
where each T
l
consists of a set of three vertices:
={
,
,
,
,
,
}
(2)
GEOMETRICAL CONSTRAINTS FOR LIGAND POSITIONING
205

Center, normal and area of each triangle T
l
, namely
g
l
,
and A
l
, respectively , can be computed by:
=(
,
+
,
+
,
)/ 3
(3)
=
,
−
,
×
,
−
,
(4)
=
,
−
,
×
,
−
,
/ 2
(5)
while the total area of the mesh A is given by
cumulating the area of each single triangle:
=
(6)
where the Gaussian sphere is partitioned into a
number of cells m.
Then all the triangle T of the target molecule are
mapped onto the corresponding cells on the basis of
the orientation
.
In the approach described in this paper it has
been adopted the EC-EGI solution. In this
framework, in the Gaussian sphere are mapped the
surface patches according to their orientation with a
weight composed of three complex numbers:
,
=
,
,
;
,
=
,
,
;
(7)
,
=
,
,
;
where
is the direction associated with a point on
the Gaussian sphere,
the total number of surface
patches with normal
,
,
the area of the lth
surface patch with normal
, and [
,
,
,
,
,
] are
the 3D coordinates of the mass center of the lth
surface patch. Note that the EC-EGI representation
can be seen as three CEGI representations, one for
each one of the main axis. Moreover, if the object is
convex the mass center of the three
,
, and
distributions on the Gaussian sphere coincides with
the center of the sphere. In fact, this is true also for
the EGI (and for the CEGI), it is:
,
=
,
=
=
,
=
=
=0
(8)
Since for convex object
,
=
,
=
,
=
, being
the area of the surface patch with
normal
.
It is also easy to show that
,
,
,
is translation invariant
(i.e. the magnitude of the EC-EGI representation is
translation invariant).
The Extended Gaussian Image does not encode
any position information; the Complex EGI encodes
the signed distance of each surface patch, and finally
the EC-EGI encodes the 3D position. With the richer
information included, the EC-EGI could remove
some of the ambiguities that CEGI has.
4 IMPLEMENTATION
A tessellated sphere with uniform, and isotropic
subdivision is needed. These properties are satisfied
by the projection of regular polyhedron onto the
sphere. Adopting the highest order regular
polyhedron, the icosahedron with twenty triangular
cells as a basis (that provides a too coarse sampling
of the orientations), and proceeding further with
precision, by dividing iteratively the triangular cells
into four smaller triangles according to the well
known geodesic dome constructions, the required
level of resolution can be achieved: being n the
number of iterative subdivision steps, the cells
number is m=10 2
2n+1
, and the area (solid angle) of
the single cells is
10 2
respectively. The
corresponding data structure is consequently a
hierarchical one (in which each cell of one level
contains, other than the specific orientation, the four
pointers to cells of the subsequent level) and
hierarchical is the searching strategy of the
orientation histogram values.
5 THE MATCHING PROBLEM
Given two candidates dual parts of proteins (i.e. a
cavity and a ligand) the aim is to find if they are
geometrically compatible, that is about findings the
rigid motion that could bring the protrusion into the
cavity. On this purpose we apply a preliminary
coarse constraint given by the mass of the EC-EGI
of the cavity and the ligand: A
cav
>A
lig
, this constraint
is not theoretically supported, but in practice works
in all the considered cases. Satisfied this constraint,
as a matching index we experimented four
parameters:
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
206

the Minkowski distance:
=
,
−
,
(9)
in for p=1 and p=2 we obtain the Manhattan and the
Euclidean distances respectively;
the Bray Curtis distance:
=
∑
,
−
,
∑
,
+
,
(10)
obviously 0
≤
B
≤
1;
the Hausdorff distance:
=
max ( ||
∀
,
−
∀
,
||,
||
∀
,
−
∀
,
|| )
(11)
the EC-EGI distance:
for a given threshold
θ
, being n the number of
triangles for which:
,
−
,
,
,
,
≥
,
,
,
=0
(12)
the distance is given by =
⁄
, i.e. the
percentage of the triangles satisfying the threshold
criteria.
For each couple candidate protein-ligand these
parameters are applied to detect the best k
candidates active sites.
6 LIGAND POSITIONING
The candidate positions of a ligand into a cavity is
determined on the basis of two steps:
- alignment of ligand and cavity;
- detection of the set V
≡
{v} for which L
v
⊆
C;
The set V can be easily obtained through the erosion
operator ∎ of mathematical morphology:
=∎ (13)
being L the structural element of the erosion.
7 RESULTS
A first experimentation of the proposed technique
has been applied to a number of proteins (e.g PDB
IDs 1KIM, 1TNL, 2OH4, 3EHY, 3L62). The
analysis has been done with a resolution of 0.25 A°,
which entails a van der Waals radius of more than
five voxels to the smallest represented atoms. The
SES is obtained from the van der Waals surface,
after the execution of a closure operator, using a
sphere with radius of 1.4 A°, approximately 6 voxels
(corresponding to the conventional size of a water
molecule), as structural element.
For what concerns the pockets detection the three
parameters quoted in (Cantoni, 2010) have been set
as follows: the minimum travel depth of the local
tops TD
LT
within a range of [25,50] voxels; the
nearness of others, more significant, local tops to
τ1=200 voxels and the relative values of the local-
top travel-distance to τ2=2000 voxels. Moreover, the
volume of the water molecule has been set to XX
voxels.
In particular we will show here the results for
protein with PDB ID 3EHY. In this case the quoted
parameter TDLT has been set to 47 voxels.
particular structure.
In figure 1 it is shown the final result of the
segmentation process of the protein 3EHY for the
detection of pockets and tunnels. Note that among
the 25 pockets that have been detected, we have
considered only the five most extensive.
Figure 1: Result of the segmentation process of PDB ID
3EHY for the detection of pockets.
Referring to computational performance, our
algorithm runs on an Intel Q6600 Processor with 4
GB of Ram. The analysis of pockets and
protuberances on 3EHY protein has been done in 45
seconds starting from the PDB file. All the matching
of the ligand with all the pockets of the protein has
been done in 125 ms.
GEOMETRICAL CONSTRAINTS FOR LIGAND POSITIONING
207
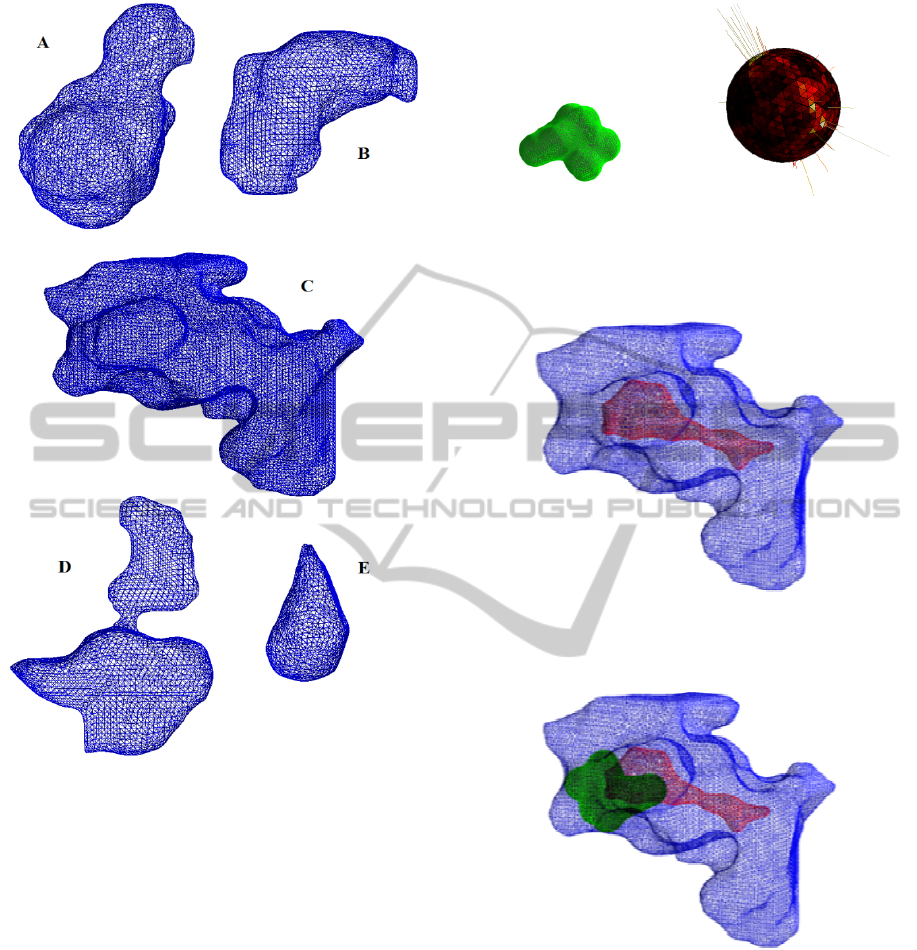
Figure 2: The five most extensive pockets for protein
3EHY.
8 CONCLUSIONS
The aim of this activity is the identification of
candidate locations for a given ligand in a given
protein. The approach is based on an evaluation
up-to-now only geometrical and topological, but we
are now working for the introduction of the
biochemical aspects.
For the morphological analysis we are
proposing the technique of the Enriched Complex
Extended Gaussian Image.
The achieved results look very promising as it
seems to improve something not only from the
computational point of view. We started an
extensive experimentation phase to validate our
solution and to identify the best practice for our new
approach.
(a)
(b)
Figure 3: Wireframe surface representation of the ligand
with ID TBL. b) The common correspondent modulus of
the three components of the CE-EGI.
Figure 4: Cavity C of the protein 3EHY in blue and the
locus of the candidates position for which the ligand TBL
is completely contained.
Figure 5: Cavity and a generic candidate position of the
ligand corresponding to the point indicated with a star in
figure 4.
REFERENCES
Bock, M.E., Garutti, C., Guerra, C., 2007. Spin image
profile: a geometric descriptor for identifying and
matching protein cavities. Proc. of CSB, San Diego.
Cantoni, V., Gatti, R., Lombardi, L., 2010. Segmentation
of SES for Protein Structure Analysis. In Proceedings
of the 1st International Conference on Bioinformatics.
BIOINFORMATICS 2011 - International Conference on Bioinformatics Models, Methods and Algorithms
208

BIOSTEC 2010. Valencia (ES). Jan 20-23, 2010, pp.
83-89.
Frome, A., Huber, D., Kolluri, R., Baulow, T., Malik, J.,
2004. Recognizing Objects in Range Data Using
Regional Point Descriptors. Computer Vision - ECCV,
(2004), pp. 224-237.
Glaser, F., Morris, R.J., Najmanovich, R.J., Laskowski,
R.A., Thornton, J.M., 2006. A Method for Localizing
Ligand Binding Pockets in Protein Structures.
PROTEINS: Structure, Function, and Bioinformatics,
62, (2006), pp. 479-488.
Horn, B.K.P., 1984. Extended Gaussian images.
Proceedings of the IEEE, 72, 1671–1686.
Hu, Z., Chung, R., Fung K. S. M. 2010. EC-EGI: enriched
complex EGI for 3D shape registration. Machine
Vision and Applications, 2, 177–188.
Kang, S.B., Ikeuchi, K., 1991. Determining 3-D object
pose using the complex extended Gaussian image.
IEEE Computer Society Conference on Computer
Vision and Pattern Recognition, pp. 580–585.
Kang, S., Ikeuchi, K., 1993. The complex EGI, a new
representation for 3D pose determination. IEEE
Trans. Pattern Anal. Mach. Intell., 15(7), 707–721.
Matsuo, H., Iwata, A., 1994. 3-D Object Recognition
Using MEGI Model from Range Data. Proc. 12th Int’l
Conf. Pattern Recognition, Jerusalem, Israel, pp. 843-
846.
Shulman-Peleg A., Nussinov, R., Wolfson, H.,
Recognition of Functional Sites in Protein Structures.
J. Mol. Biol., 339, (2004), pp. 607-633.
Wang, D., Zhang, J., Wong H.S., Li, Y., 2007. 3D Model
Retrieval Based on Multi-Shell Extended Gaussian. G.
Qiu et al. (Eds.): VISUAL 2007, LNCS 4781, Springer-
Verlag Berlin Heidelberg, pp. 426–437.
Zhang, J., Wong H.S., Yu, Z., 2006. 3D Model Retrieval
Based on Volumetric Extended Gaussian Image and
Hierarchical Self Organizing Map. MM’06, October
23–27, 2006, Santa Barbara, California, USA., ACM
1-59593-447-2/06/0010, 121-124.
GEOMETRICAL CONSTRAINTS FOR LIGAND POSITIONING
209
