
NUCLEAR MEDICINE IMAGE MANAGEMENT SYSTEM
FOR STORAGE AND SHARING BY USING GRID SERVICES
AND SEMANTIC WEB
Daniela Giordano, Carmelo Pino, Concetto Spampinato
Department of Informatics and Telecommunication Engineering, University of Catania, 95125 Catania, Italy
Marco Fargetta
Italian Institute for Nuclear Physics (INFN), Catania, Italy
Angela Di Stefano
Institute of Neurological Science, Italian Research Council (CNR), Catania, Italy
Keywords:
Medical data sharing, Logical file catalogue (LFC), Metadata service (AMGA), SPECT, PET, Web 2.0.
Abstract:
Large amounts of images (SPECT, PET, scintigraphy) in the nuclear medicine field have been routinely
produced in the last decades. In this paper we propose an image management system that allows nuclear
medicine physicians to share the acquired images and the associated metadata both locally (i.e. within the
same medical institute) and globally with other nuclear medicine physicians located anywhere in the world
by using GRID services for data (LFC) and metadata (AMGA) storage. The proposed system guarantees
medical data protection by anonymization that removes most sensitive data for unauthorized users, and
encryption, that guarantees data protection when it is stored at remote sites. Another important issue is that
often nuclear medicine data is associated with other medical data (e.g. neurological data) for diagnosis and
therapy follow-up. In order to correlate images with other clinical information, the common metadata are
enriched by developing a controlled vocabulary, which integrates known standards such as FOAF, CCR and
GeneOntology. All the metadata are stored in an RDF (Resource Description Framework) repository in order
to make the system fully compatible with existing metadata storage systems following the semantic web’s
philosophy.
1 INTRODUCTION
Usually, nuclear medicine physicians carry out ex-
aminations, e.g. Single Photon Emission Computed
Tomography (SPECT), Positron Emission Tomogra-
phy (PET) or scintigraphy, without a complete knowl-
edge of the clinical history of the patients. In fact,
most of the raw data collected in experiments or clin-
ical trials is usually stored either in different places
or in separate paper forms. Therefore there is a real
need of integrating heterogeneous data sources in or-
der to have a holistic vision of the patient’s health sta-
tus. Currently, existing methods, e.g. (Gabber et al.,
2003), (Xu and Jiang, 2009) barely address medical
data management needs beyond the specific depart-
ment’s boundaries, while it is known that the patient
medical folders are wide spread over many medical
sites involved in the patient’s healthcare. For this rea-
son, many efforts have been done in last years to de-
velop an interoperable infrastructure for digital health
((Cheung et al., 2009), (Freund, 2006)) by using se-
mantic web concepts, even if the attention is mainly
oriented to metadata sharing. A relevant project in
this direction is the e-Child project (Freund, 2006)
that develops a healthcare platform for pediatrics and
aims at integrating ontologies to homogenize biomed-
ical (from genomic, through cellular, disease, patient
and population-related) data. The system proposed in
this paper follows, and for some aspects, overcomes
the above medical data management’s approach be-
80
Giordano D., Pino C., Spampinato C., Fargetta M. and Di Stefano A..
NUCLEAR MEDICINE IMAGE MANAGEMENT SYSTEM FOR STORAGE AND SHARING BY USING GRID SERVICES AND SEMANTIC WEB.
DOI: 10.5220/0003170500800086
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2011), pages 80-86
ISBN: 978-989-8425-34-8
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

cause it permits to store images associating them with
not directly related metadata (i.e. not included in the
DICOM file) by developing an RDF controlled vocab-
ulary, which links the most common ontologies used
in the medical field with personal health records on
the Internet (e.g. Google Health). This makes our
system interoperable with methods that share data fol-
lowing semantic web (e.g. in (Holford et al., 2009))
enriches the common information with personal data,
i.e. not present in medical facilities.
Given the huge amount of medical data involved in
this process and the need of collaboration among re-
searchers our system is provided with a GRID layer.
Many GRID systems have been proposed for han-
dling medical data geographically distributed on var-
ious medical centers, such as HOPE (Diarena et al.,
2008), MediGrid (Montagnat et al., 2005) or MAM-
MOGRID (Amendolia, 2004), but most of them deal
only with data files and do not provide higher level
services for manipulating medical data or for handling
the associated metadata, and also lack in the integra-
tion with other systems.
Therefore, in this paper we propose an approach for
sharing and analyzing nuclear medicine images where
1) metadata are stored in RDF, thus giving the possi-
bility to easily grab information coming from loca-
tions who share data according to the web semantic
approach and 2) image sharing among different med-
ical institutes is handled by GRID services in a trans-
parent way for the users. The key features of the pro-
posed system are:
• Full functionality in cases of network malfunc-
tioning is ensured since data are stored both lo-
cally (in the user’s computer and in the medical
institute server) and globally on GRID;
• It is interoperable with existing sharing methods
that use semantic web;
• It is flexible since researchers can share content
choosing when and what to share;
• The metadata, usually contained in the DICOM
files, are enriched in order to take into account the
whole clinical history of patients. Users may de-
cide to export all the information (data and meta-
data), only data, only metadata or only part of it;
• Data encryption is implemented using SSH,
which ensures data integrity over Internet trans-
fers;
• Data privacy is guaranteed by removing sensitive
information before any transmission.
The paper is so organized: in section 2 the storage
and sharing system is described. In detail, we present
how the storage and the sharing work inside a medi-
cal institute using a client-server architecture and how
it works among different institutes using GRID ser-
vices. Sect. 3 and Sect. 4 describes, respectively, the
high level features for managing the interoperability
with other systems, for image analysis and for system
querying and the user interface. Finally, concluding
remarks are given.
2 STORAGE AND SHARING
SYSTEM OVERVIEW
In order to develop a distributed environment for im-
age and information sharing to support the diagnosis,
the treatment of patients and for statistical evaluation,
the system is provided with two levels of storage and
sharing: the first is locally managed by a client-server
architecture, deployed in the medical institute where
the nuclear medicine physicians belong to, whereas
the second one is on GRID and allows global data
sharing, i.e. data may be shared among different in-
stitutes using the services offered by the GRID com-
puting. Fig. 1 shows the architecture of the proposed
system for the local and global data sharing.
The typical use case is the following: a nuclear
medicine physician stores the images and the meta-
data of a performed examination in its own local
database (located in his/her computer). Afterwards,
the client creates an anonymous version of the data re-
moving all the confidential information so they can be
sent to the main server in the respect of privacy issues.
Additionally, the client allows users to define the set
of metadata he/she wants to share both in GRID and
in his/her medical institute.
The data transmission between client and server runs
asynchronously in order 1) to make the system robust,
in fact in case that, Internet connection is unavailable,
data are locally stored and subsequently sent to the
main server and to GRID when the connection will
be available again and 2) to keep users unaffected by
the actual time needed for the data transfer.
The server is provided with repositories where all
the metadata produced within the same institute are
stored. The communication with GRID is delegated
to the server, thus optimizing the bandwidth’s use.
2.1 Local Data Storage and Sharing
Inside a medical institute, data are stored and shared
using a standard client-server architecture, as shown
in fig. 2. The client and the server are connected by a
local network or a VPN (Virtual Private Network).
The client contains the user interfaces and implements
the logical communication with the GRID infrastruc-
ture. It also contains a file repository (for image stor-
NUCLEAR MEDICINE IMAGE MANAGEMENT SYSTEM FOR STORAGE AND SHARING BY USING GRID
SERVICES AND SEMANTIC WEB
81
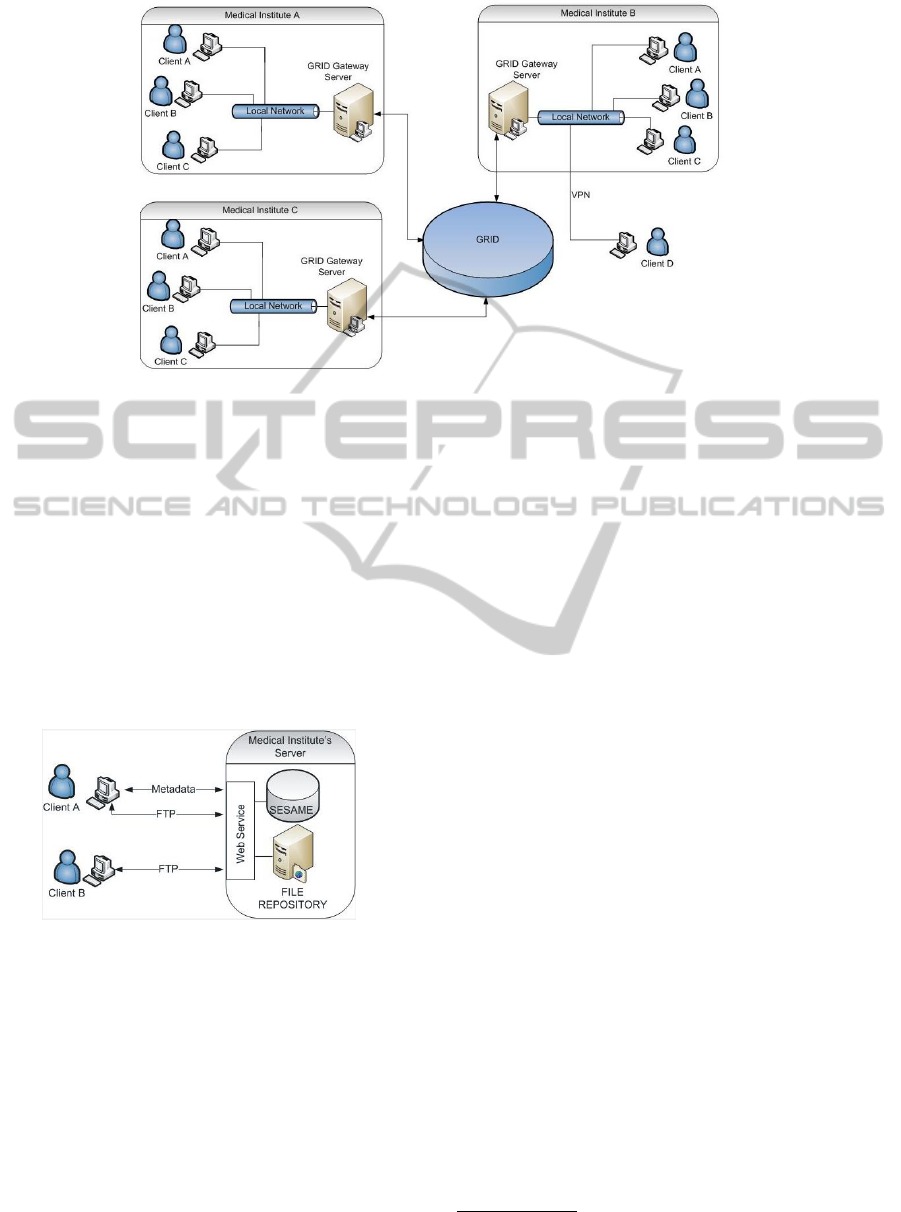
Figure 1: Architecture for Local and Global Data Storage and Sharing.
age) and a SESAME server (Broekstra et al., 2001)
(for RDF metadata storage), in order to save patient’s
data locally.
The server also includes a file repository and a
SESAME metadata repository for the data produced
by all the nuclear medicine physicians within the
same institute. Data are sent from the client to the
server using File Transfer Protocol (FTP), whereas
metadata is transmitted to SESAME server using
Simple Object Access Protocol (SOAP) requests by
means of a webservice, as shown in fig. 2.
Figure 2: Local Data Storage and Sharing.
By using the client interface, a nuclear medicine
physician can record and manage patients, add infor-
mation to patient’s clinical history (according to the
schema shown in the next section), include any rele-
vant documents (textual reports, generic images, DI-
COM images, etc..), run queries locally or on GRID
data, associate the metadata deriving from the queries
to the data locally stored and perform statistical anal-
ysis on sets of data and virtual data (i.e. coming from
the main institute center or from GRID).
2.2 Global Data Sharing on GRID using
LFC and AMGA
Data sharing among different institutes, geograph-
ically distributed, is implemented on GRID and it
is based on the paradigm to create virtual environ-
ments where large amount of data and complex com-
putations can be performed by different communi-
ties grouped in Virtual Organisation (VO). Usually,
each VO offers several services for GRID participants
in order to simplify the data management providing
them basic functionality to store and retrieve files.
The access to the GRID is hidden by a middleware.
In this work we used the EGEE Grid and the G-Lite
middleware
1
. The two main services of the G-lite
middleware for data and metadata storage, used in
our system, are: the Logical File Catalogue (LFC)
(Venugopal et al., 2004) and the AMGA Metadata
Service (Nuno, 2006). The LFC allows users to as-
sociate a logical name to a file in a hierarchy for-
mat like a local file system, hiding the real location
of the storage. Moreover, a logical name may refer
many replicas, so a user can retrieve the file through
its logical name from the nearest location in a trans-
parent way. The AMGA Metadata Service is a special
database designed to store metadata associated with
files. Therefore, its internal structure reproduces a file
system hierarchy where the directories are collections
of metadata defined in a custom schema and each file
in the directory is an entry containing the values for
the metadata. This approach allows to easily map a
file name with a set of metadata inside AMGA.
1
EGEE website - http://www.eu-egee.org/ and gLite
Grid middleware website -http://www.glite.org/
HEALTHINF 2011 - International Conference on Health Informatics
82
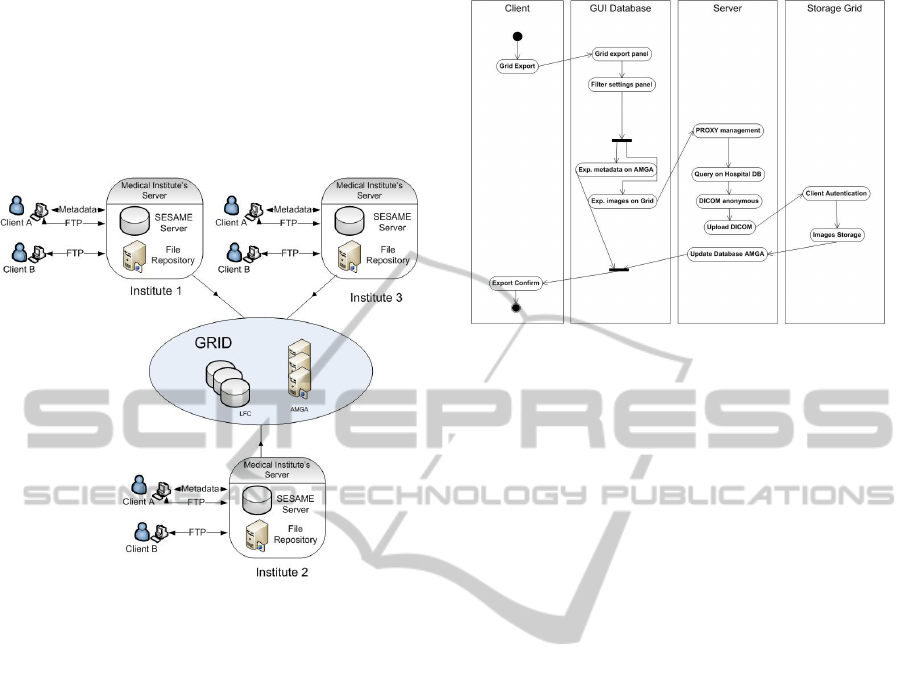
The communication with GRID infrastructure is man-
aged by the medical institute server, which also aims
at maintaining aligned the information stored in nu-
clear medicine physicians’ computers and the one
stored in GRID. In each institute the main server
represents the bridge between clients and GRID, as
shown in fig. 3.
Figure 3: Global Data Storage and Sharing.
The flow diagram of the interaction between a
generic medical institute and the GRID infrastructure
is shown in fig. 4, when a nuclear medicine physician
requires the storage of a specific image on GRID by
using a proper GUI (Graphical User Interface). The
steps performed by our system are: 1) the client sends
to the server the proxy certificate (needed for the ac-
cess to GRID) previously created and the identifier of
the image to be stored on GRID, 2) the server of the
medical institute (for simplicity called proxy server)
queries the file repository to check if the file is present
or not, if not it asks the client to send the image, then
it sets the necessary permissions to read, write for the
GRID, 3) the server queries the GRID database using
the medical institute Grid ID given by the client in or-
der to see if the image was already stored, if not 4) the
server removes sensitive data, sends it to a Grid Stor-
age Element, writes in the LFC catalog an appropri-
ate logical name and writes (uploads if the image was
previously stored) the metadata in the AMGA server.
3 HIGH LEVEL FEATURES
In order to provide useful information about the stored
images and to make them available with the related
Figure 4: Interaction with Medical Institute - GRID Infras-
tructure.
metadata to the nuclear medicine community, the sys-
tem is provided with high level features. More in de-
tail, the system contains three processing levels:
• A semantic layer that enriches patient metadata
by a controlled vocabulary using RDF/XML stan-
dard. This level guarantees the interoperability
with existing frameworks;
• An image processing layer that analyzes the
stored images. This is an important layer, since
sometimes is desirable to share only the process-
ing results and not the entire image. This level
performs the image analysis and interacts with the
semantic layer for the storage of the processing re-
sults in RDF/XML;
• Query Composition for performing complex
queries both locally and on GRID. This module
allows users to search useful information by pro-
cessing only the metadata available locally or in
GRID.
The interaction between the three levels and the
system’s architecture is shown in fig. 5.
3.1 Semantic Layer
Usually nuclear medicine images (e.g. in PET,
SPECT) are stored in DICOM format, containing the
metadata provided with the standard. These metadata
are not sufficient for describing the clinical history
of patients. For this reason we enrich the informa-
tion available in order to give the nuclear medicine
physicians the possibility to better figure out a spe-
cific disease by developing a model that represents
the medical data so that it can be analyzed by seman-
tic tools. In detail, the system stores concepts, speci-
fies typed relationships between these concepts using
NUCLEAR MEDICINE IMAGE MANAGEMENT SYSTEM FOR STORAGE AND SHARING BY USING GRID
SERVICES AND SEMANTIC WEB
83
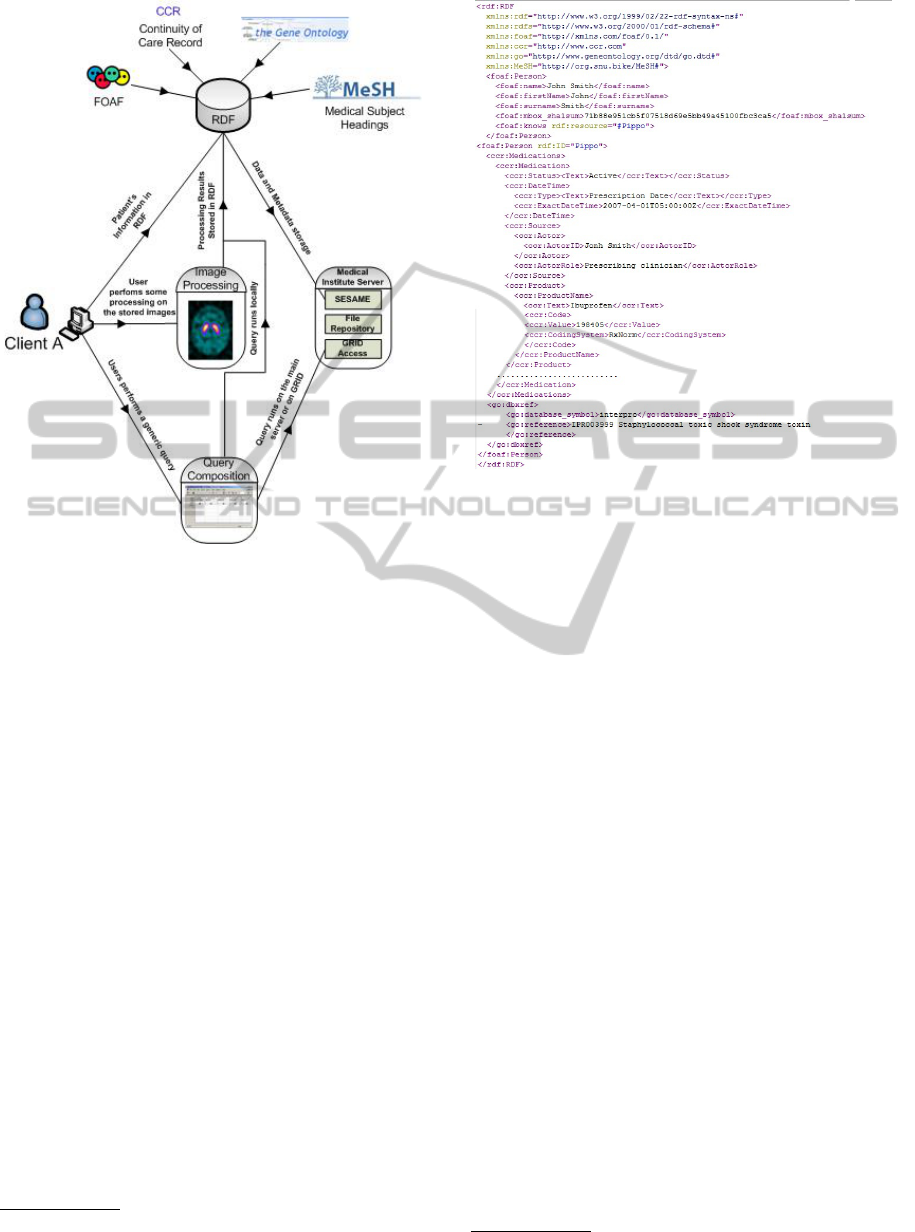
Figure 5: Interaction between the high level features’ sys-
tems.
RDF
2
(Resource Description Framework) with XML
syntax format. More in detail, we enrich the DICOM
metadata by developing a controlled vocabulary that
includes:
• Personal data by using FOAF ontology
3
;
• Generic Health Information according to the Con-
tinuity of Care Record (CCR) standard (Detmer
et al., 2008) such as: diagnoses, allergies, medi-
cation list, immunizations, family history, social
history, vital signs, procedures, symptoms, plan
of care, functional status, biosignals (EEG, ECG,
etc...);
• Genetic information using GeneOntology (Ash-
burner, 2000);
• Neurological detailed information by using Mesh
(Soualmia et al., 2004);
• Image processing information that represents the
output of the implemented image processing algo-
rithms and which introduces a new semantic level
to the stored metadata.
An example of RDF file for a patient is shown
in fig. 6. It is notable that this information is in-
serted by the users, but it can be easily obtained by
2
http://www.w3.org/RDF/
3
http://www.foaf-project.org/
Figure 6: Example of produced RDF File.
querying systems that share data using RDF. For in-
stance, personal data in FOAF can be derived from a
generic social network or by using a v-card, whereas
generic health information can be obtained by the
user’s Google Health Account
4
or other systems that
aim at storing online health care data such as the one
proposed in (Bielikov
´
a and Moravc
´
ık, 2008).
Metadata storage has been carried out by using
SESAME server (Broekstra et al., 2001) so that these
information may be available also for other purposes.
The sensitive data, such as Name, Surname, SSN
must be available only for the nuclear medicine physi-
cian who carries out the examination, and are not ex-
ported in RDF in order to ensure data privacy.
3.2 Image Processing Layer
This level is provided with a set of processing meth-
ods for medical image analysis. The output of this
processing is stored according to the semantic layer
and is related to the specific processed image. This
allows users to also share the results of the processing
avoiding to send the original images when it is not re-
quired, resulting in less bandwidth usage.
The implemented methods for nuclear image analy-
sis, also available for MRI, X-Rays images, are:
• Measurement of distances, angles and some pa-
rameters within the images;
• The contrast absorption curve over time;
4
https://www.google.com/health/
HEALTHINF 2011 - International Conference on Health Informatics
84
• Image Texture and Image Contour Analysis for
specific organs;
• Pattern recognition for identifying brain struc-
tures.
Example of such algorithms have been proposed
by the authors in (Faro et al., 2010a), (Faro et al.,
2010b) and (Giordano et al., 2009). Therefore, when
a user performs one of the above methods, the out-
put is treated as metadata and stored in the SESAME
server.
3.3 Query Composition
The query composition level aims at building com-
plex queries both locally and on GRID. The queries
are performed only on the metadata (stored in the
SESAME server and in the AMGA server) since a
content based image retrieval module is not present.
This level receives the user’s queries (by using a con-
trolled GUI) and interacts both with the local storage,
performing SPARQL queries on the SESAME server,
and with the GRID, where queries are performed fol-
lowing the approach proposed in (Montagnat et al.,
2008).
4 CLIENT INTERFACE
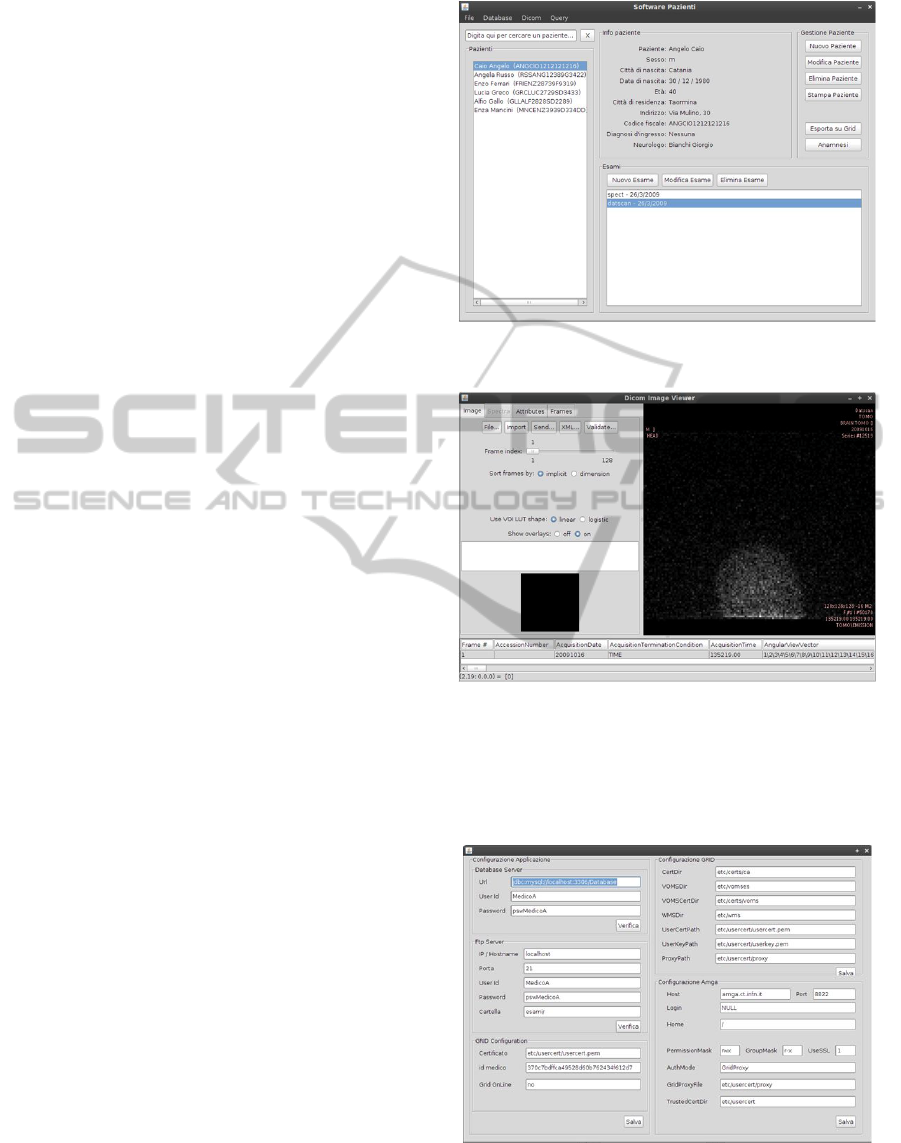
The client interface consists of a multiform Java ap-
plication, allowing users to manage any type of infor-
mation concerning the patients. The functionalities
available for nuclear medicine physicians are:
• view information about their patients ;
• search a patient by typing the name;
• insert, modify or delete a patient;
• print a patient’s report;
• show the patient exams;
• insert, modify or delete an exam;
• management of medical history;
• view and process DICOM files;
• export the patient’s data on GRID;
• query the system both locally and globally on
GRID.
For each functionality a specific GUI has been de-
signed. For example, fig. 7 shows the main interface
where a list of patients and a tool to manage patients’
data is available. In the bottom area, there is also a
section to handle the exams for each patient. Fig. 8,
instead, shows the interface for DICOM files and im-
age processing.
Figure 7: Main GUI.
Figure 8: GUI for DICOM File Processing.
Moreover, for expert users the client application
contains a form (fig. 9) to set parameters for the FTP,
GRID and AMGA.
Figure 9: GUI for parameters’ setting.
NUCLEAR MEDICINE IMAGE MANAGEMENT SYSTEM FOR STORAGE AND SHARING BY USING GRID
SERVICES AND SEMANTIC WEB
85

5 CONCLUDING REMARKS
In this paper we presented a distributed system of
data and metadata storage, sharing and processing.
Web semantic data modeling using RDF permits that
all the information can be stored and reused. The
RDF modeling allows us to efficiently integrate het-
erogeneous data into a coherent whole and to provide
description of data elements. The area of applica-
tion, is still confined to the specific sector of nuclear
medicine but the flexibility of the architecture permits
the application to be used on a greater number of med-
ical fields by a simple re-modeling. The platform will
be integrated with an image retrieval system in order
to assist the physicians in the retrieval of images by
content. A future development is to provide the sys-
tem with an ontology layer that will map high level
requests to low level queries.
REFERENCES
Amendolia, S. R. (2004). Mammogrid: A service oriented
architecture based medical grid application. In GCC,
pages 939–942.
Ashburner, M. (2000). Gene ontology: Tool for the unifica-
tion of biology. Nature Genetics, 25:25–29.
Bielikov
´
a, M. and Moravc
´
ık, M. (2008). Modeling the
reusable content of adaptive web-based applications
using an ontology. In Advances in Semantic Media
Adaptation and Personalization, pages 307–327.
Broekstra, J., Kampman, A., and Harmelen, F. V. (2001).
Sesame: An architecture for storing and querying rdf
data and schema information. In Semantics for the
WWW. MIT Press.
Cheung, K. H., Prud’hommeaux, E., Wang, Y., and
Stephens, S. (2009). Semantic Web for Health Care
and Life Sciences: a review of the state of the art.
Brief. Bioinformatics, 10:111–113.
Detmer, D., Bloomrosen, M., Raymond, B., and Tang, P.
(2008). Integrated personal health records: Transfor-
mative tools for consumer-centric care. BMC Medical
Informatics and Decision Making, 8(1):45.
Diarena, M., Nowak, S., Boire, J. Y., Bloch, V., Donnar-
ieix, D., Fessy, A., Grenier, B., Irrthum, B., Legre,
Y., Maigne, L., Salzemann, J., Thiam, C., Spalinger,
N., Verhaeghe, N., de Vlieger, P., and Breton, V.
(2008). HOPE, an open platform for medical data
management on the grid. Stud Health Technol Inform,
138:34–48.
Faro, A., Giordano, D., Pino, C., Spampinato, C., and
Di Stefano, A. (2010a). 3D striatum reconstruction
of 123ioflupane MRI images for quantitative assess-
ments on the dopaminergic neurotransmission system.
In IEEE International Workshop on Medical Mea-
surements and Applications (MeMeA 2010), Ottawa,
Canada.
Faro, A., Giordano, D., Spampinato, C., and Pennisi, M.
(2010b). Statistical texture analysis of MRI images to
classify patients affected by multiple sclerosis. In 12th
Mediterranean Conference on Medical and Biological
Engineering and Computing MEDICON 2010, pages
236–239, Porto Carras, Chalkidiki, Greece. Interna-
tional Proceedings of the IFBME, Springer.
Freund, J. (2006). Health-e-child: an integrated biomedical
platform for grid-based paediatric applications. Stud
Health Technol Inform, 120:259–270.
Gabber, E., Fellin, J., Flaster, M., Gu, F., Hillyer, B.,
Ng, W. T.,
¨
Ozden, B., and Shriver, E. A. M. (2003).
Starfish: highly-available block storage. In USENIX
Annual Technical Conference, FREENIX Track, pages
151–163.
Giordano, D., Scarciofalo, G., Spampinato, C., and
Leonardi, R. (2009). Automatic skeletal bone age as-
sessment by integrating EMROI and CROI process-
ing. In IEEE International Workshop on Medical
Measurements and Applications (MeMeA 2009), Ce-
traro, Italy.
Holford, M. E., Rajeevan, H., Zhao, H., Kidd, K. K., and
Cheung, K. H. (2009). Semantic Web-based inte-
gration of cancer pathways and allele frequency data.
Cancer Inform, 8:19–30.
Montagnat, J., Breton, V., and Magnin, I. (2005). Partition-
ing medical image databases for content-based queries
on a Grid. Methods Inf Med, 44:154–160.
Montagnat, J., Frohner,
´
A., Jouvenot, D., Pera, C., Kun-
szt, P., Koblitz, B., Santos, N., Loomis, C., Texier, R.,
Lingrand, D., Guio, P., Rocha, R. B. D., de Almeida,
A. S., and Farkas, Z. (2008). A secure grid medical
data manager interfaced to the glite middleware. J.
Grid Comput., 6(1):45–59.
Nuno, N. S. (2006). Distributed metadata with the amga
metadata catalog. In Workshop on NextGeneration
Distributed Data Management - HPDC-15.
Soualmia, L., Golbreich, C., and Darmoni, S. (2004). Rep-
resenting the mesh in owl: Towards a semi-automatic
migration. In Proceedings of the KR 2004 Work-
shop on Formal Biomedical Knowledge Representa-
tion, pages 81–87.
Venugopal, S., Buyya, R., and Winton, L. (2004). A grid
service broker for scheduling distributed data-oriented
applications on global grids. In MGC ’04: Proceed-
ings of the 2nd workshop on Middleware for grid com-
puting, pages 75–80, New York, NY, USA. ACM.
Xu, Z. and Jiang, H. (2009). Hass: Highly available, scal-
able and secure distributed data storage systems. Com-
putational Science and Engineering, IEEE Interna-
tional Conference on, 2:772–780.
HEALTHINF 2011 - International Conference on Health Informatics
86
