
A MULTI-AGENT TOOL TO ANNOTATE BIOLOGICAL
SEQUENCES
C´elia Ghedini Ralha, Hugo Wruck Schneider, Maria Emilia M. T. Walter
Department of Computer Science, University of Bras´ılia, Campus Universit´ario Darcy Ribeiro
PO Box 4466, Bras´ılia, ZipCode 70.904-970, Brazil
Marcelo M. Br´ıgido
Institute of Biology, University of Bras´ılia, Campus Universit´ario Darcy Ribeiro
Bras´ılia, ZipCode 70.910-900, Brazil
Keywords:
Multi-agent system, Annotation, Genome sequencing projects, BioAgents.
Abstract:
Nowadays, great challenges are imposed by the existence of enormous volume of DNA and RNA sequences,
which are continuously being discovered by genome sequencing projects, through the automatic sequencers
based on massively parallel sequencing technologies. Thus, the task of identifying biological function for
these sequences is a key activity in these high-throuput sequencing projects, where the automatic annotation
must be significantly improved. In this context, this paper presents a multi-agent approach to address the
important issue of automatic annotation in genome projects. We developed a sophisticated prototype named
BioAgents, which simulates biologists knowledge and experience to annotate DNA or RNA sequences in
genome sequencing projects, where different specialized intelligent agents work together to accomplish the
annotation process.
1 INTRODUCTION
Artificial Intelligence (AI) techniques are being em-
ployed in bioinformatics with increasing success. In
this context, a MAS takes goal-oriented approach in
which agents can act cooperatively to reach a goal.
Recently, new massively parallel sequencing tech-
nologies (Mardis, 2008), that can produce billions of
bases in a veryshort time, havedramatically increased
the amount of biological data, and have presented new
bioinformatics challenges (Pop and Salzberg, 2008).
A key activity in these genome projects is the annota-
tion of these enormous volume of sequences.
The annotation phase of a genome project has
the objective of assigning biological functions to the
identified DNA and RNA sequences. In addition,
ab initio gene finding programs (programs that find
genes based on biological and chemical properties)
to predict protein-coding genes are being largely em-
ployed, using annotation techniques that include com-
parisons among the investigated sequences and se-
quences of related species, available in public biolog-
ical data bases, like GenBank (Benson et al., 2008).
In the annotation phase, computational methods
to infer biological functions to each sequence are nor-
mally accomplished by approximate string matching
algorithms, like BLAST (Altschul et al., 1990). These
algorithms run on data bases containing the sequences
and their already identified functions, or methods to
identify noncoding RNAs (Eddy and Durbin, 1994).
The annotation process can be completed by biolo-
gists, who using their knowledge analyze and correct
the function suggested by the programs.
The annotation phase in the context of massively
parallel DNA pyrosequencing could be certainly im-
proved by computational tools. Thus, this paper fo-
cuses on the use of AI techniques to bioinformatics,
with a MAS approach implemented in a prototype
called BioAgents. This version of BioAgents, has a
Web interface, uses BLAST and BLAT (Kent, 2002)
algorithms, a method to identify noncoding RNAs –
PORTRAIT (Arrial et al., 2007), and an open source
rule engine in Java – Drools.
The rest of this work is divided into five sections.
In Section 2, we discuss previous work; while in Sec-
tion 3, the related work. In Section 4, we present the
architecture, BioAgents new prototype and MAS fea-
tures. In Section 5, we discuss the experiments. Fi-
226
Ghedini Ralha C., Wruck Schneider H., Emilia M. T. Walter M. and M. Brígido M..
A MULTI-AGENT TOOL TO ANNOTATE BIOLOGICAL SEQUENCES.
DOI: 10.5220/0003179102260231
In Proceedings of the 3rd International Conference on Agents and Artificial Intelligence (ICAART-2011), pages 226-231
ISBN: 978-989-8425-41-6
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

nally, in Section 6, we conclude and suggest future
work.
2 PREVIOUS WORK
We have already presented the first prototype of
BioAgents (Lima, 2007). It was a tool to be used with
the traditional Sanger technology, with the objective
to assist the biologists during the manual annotation
phase on genome sequencing projects. BioAgents
was developed with a three layer architecture using
JADE framework (Bellifemine et al., 2007), compar-
ing algorithms such as BLAST and FASTA (Pearson
and Lipman, 1988), using Jess inference engine (Hill,
2003).
During this first stage of our research project, the
MAS approach has proved to allow the interaction
of specialized software agents in the reach of an ob-
jective (Weiss, 2000; Wooldridge, 2009). Different
agents using specific algorithms, which interact to
each other in order to reach a common objective, have
accomplished well the process of annotation. Thus,
BioAgents was used for supporting manual annota-
tion on three different genome sequencing projects:
Paracoccidioides brasilienses fungus, Paullinia cu-
pana (guaran´a) plant and Anaplasma marginale rick-
ettsia. The obtained results were encouraging at the
traditional Sanger technology (Ralha et al., 2008).
Considering an ongoing research work, BioA-
gents was described to illustrate the recent research
field of Agent-Mining Interaction and Integration
(AMII) (Ralha, 2009). In another perspective, we
have implemented a reinforcement learning (RL)
method to BioAgents, where we have used together
with the genome sequencing projects previously ex-
perimented – Paracoccidioides brasiliensis (Pb fun-
gus) and Paullinia cupana (Guaran´a plant), together
with two reference genomes – Caenorhabditis ele-
gans and Arabidopsis thaliana, respectively, for Pb
and Guaran´a. The results obtained with the learning
layer were better when compared to the system with-
out the proposed method (Ralha et al., 2010).
3 RELATED WORK
Bioinformatics is a research area concerned with the
investigation of tools and techniques from computer
science to solve problems from molecular biology
(see Setubal and Meidanis (1997) for details). Many
projects on bioinformatics use AI techniques on dif-
ferent bioinformatics tasks such as analysis and pre-
diction of gene function. We cite some of these initia-
tives, but not being exhaustive.
Another MAS tool is the MASKS environ-
ment (Schroeder and Bazzan, 2002), that improves
symbolic learning through knowledge exchange. The
motivation is to mimic human interaction in order to
reach better solutions to data classification. The tool
Agent-based environmenT for aUtomatiC annotation
of Genomes - ATUCG is based on an agent architec-
ture, and aims to support the biologists by using the
concept of re-annotation (do Nascimento and Bazzan,
2004).
Finally, transcriptome and regulome sequenc-
ing projects, as well as metagenomics projects, se-
quenced with massively parallel sequencing tech-
nologies, have been successfully annotated with tra-
ditional annotation with Sanger technology, being
slightly modified to adequately treat the enourmous
volumes of output of these sequencers (Moore1 et al.,
2006; Solda et al., 2009; Iacono et al., 2008; Wang
et al., 2009).
Comparing to the related works cited, BioAgents
simulates biologists knowledge and experience to an-
notate DNA or RNA sequences in different genome
sequencing projects. See Section 5, where we present
our experiments conducted with a fungus, a plant and
a rickettsia project. The annotation tools cited focus
specific annotation organisms such as virus (BioMAS,
DECAF). The ATUCG focus the re-annotation pro-
cess in the traditional annotation form, while MASKS
knowledge approach is an interesting initiative, since
data classification is still an increasing problem to
genome annotation methods with massively parallel
sequencing technologies; which we believe can be
improved through the use of DM and ML techniques
(see Section 6 of future work to BioAgents).
To conclude, annotation methods for genomes
will pursue reasonably accuracy for genes presented
in other species, since sequence comparison methods
can deal well with errors, even if the genes are frag-
mented. But genes that belong uniquely to an organ-
ism will be difficult to be annotated with traditional
annotation methods, and the small size of sequences
assemblies of massively parallel sequencing projects
will increase this problem (Pop and Salzberg, 2008).
4 BIOAGENTS
Considering the new scenario of massively parallel
automatic sequencers, where billions of little frag-
ments are produced in an increasing velocity, BioA-
gents can strongly help to improve the quality of the
annotation process with its reasoning mechanism to
A MULTI-AGENT TOOL TO ANNOTATE BIOLOGICAL SEQUENCES
227
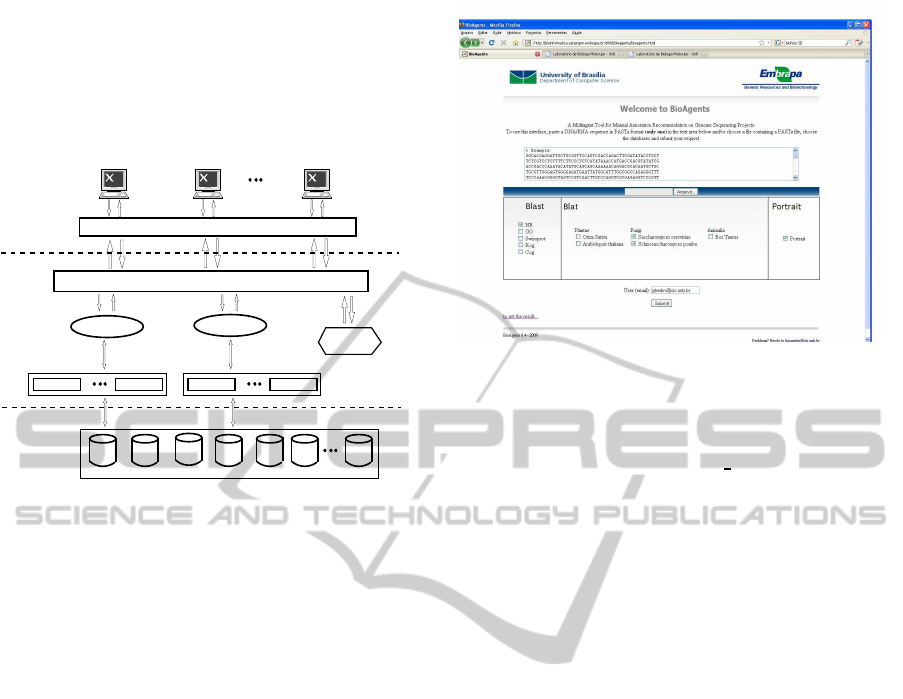
gene annotation. Figure 1 presents the architecture
of BioAgents, considering the Web prototype version,
which is divided into three layers: interface, collab-
orative and physical. Since the three layers and the
agents do not differ from previous publications, for a
more detailed explanation see (Ralha et al., 2008).
Collaborative
Layer
Interface
Layer
Interface Module
Queries Results
User 2
Queries Results
User n
Conflict Resolution Agent (CR)
Blast MR Agent Blat MR Agent
Portrait Specific
Agent (SA)
ANL Agent ANL Agent
Physical
Layer
ANL Agent ANL Agent
SC
COG
KOG
SP
GO
Swiss−
Prot
other
data
bases
nr
User 1
Queries Results
Figure 1: The three layer architecture of BioAgents.
The collaborative layer is the architecture core.
It has specialized manager agents to execute par-
ticular algorithms, like BLAST and BLAT, that in-
teract with analyst agents for treating data bases,
like nr-GenBank or kog. We defined special-
ized agents to deal with different algorithms and
specific data bases. Finally, this layer sug-
gests annotations to be sent to the interface layer
through the conflict resolution agent. The physical
layer is formed by different public biological data
bases. Figure 2 presents the BioAgents Web inter-
face (http://bioinformatica.cenargen.embrapa.br:8080/
bioagents/bioagents.html).
4.1 The Prototype
BioAgents Web prototype version was rewritten to be
time efficient in execution by using separated threads.
It was implemented with a framework for MAS devel-
opment known as Java Agent DEvelopment Frame-
work - JADE, version 3.6.1. JADE uses Java lan-
guage, and Eclipse SDK, version 3.4.1, was used as
the development environment.
JADE offers class libraries of pattern interaction
protocols, ready to be used and extended. As its plat-
form is ready to use, it is not necessary to imple-
ment agents functionalities, agent management on-
tologies and transportation mechanism for message
parsing. We have used FIPA Agent Communication
Language - FIPA ACL for message interchange and
contract net interaction protocol. BioAgents parsers
Figure 2: BioAgents Web interface.
used by the ANL agents were implemented using
some libraries of the framework BioJava, version
1.6 (http://biojava.org/wiki/Main Page). BioJava of-
fers objects to manipulate biological sequences and
parsers to files of biological sequences, among other
functionalities.
BioAgents uses Drools as a production rule sys-
tem (http://www.jboss.org/drools/). Drools is an
open source rule engine implementation written in
Java, and it is based on Charles Forgy’s Rete al-
gorithm (Forgy and Shepard, 1987) tailored for the
Java language. Drools allows pluggable language im-
plementations. With Drools we defined the biolo-
gists knowledge through the use of production rules
(declarative rules), according to the parameters de-
fined for the specific genome project.
To analyze the outputs from BLAST and BLAT,
MR and ANL agents used two parameters, the e-value
and score, according to the following rules: (i) Ver-
ify if there are alignments having e-value less than or
equal to 10
−5
(value adopted by the biologists on the
three genome projects of our experiments, but this is
a parameter of the system easily changeable);(ii) Se-
lect the lower e-value, among the alignments present-
ing the previous restriction; (iii) Select the alignment
with the higher score, if the e-values are equal.
Figure 3 presents the conflict resolution flow used
by the CR Agent. The CR agent uses BLAST and
BLAT results if at least one of them finds an e-
value less than or equal to 10
−5
. Otherwise, the CR
agent calls the PORTRAIT agent to identify noncod-
ing RNAs. PORTRAIT is a method to identify non-
coding RNA in transcriptomes of poorly character-
ized species (Arrial et al., 2007).
MR and ANL agents interpretthe results produced
by the comparison algorithms, according to the agents
knowledge formalized by production rules (presented
ICAART 2011 - 3rd International Conference on Agents and Artificial Intelligence
228
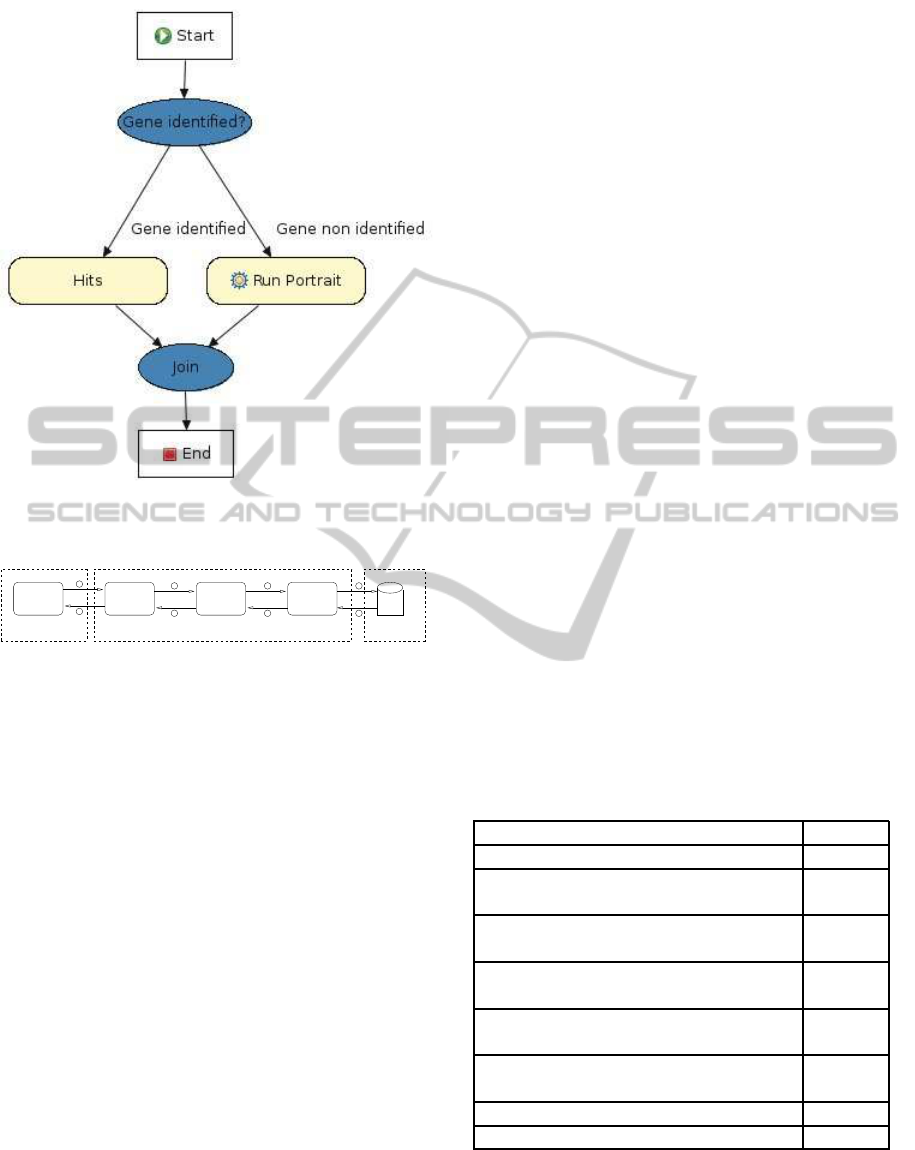
Figure 3: The set of Drools rules to analyze the outputs of
BLAST, BLAT and PORTRAIT.
Submission
Sequence
Interface Layer
Conflict
Resolution
Agent (CR)
Blast
MR Agent
ANL
Agent
nr
Physical Layer
1
2 3 4
567
8
Collaborative Layer
Figure 4: BioAgents workflow, noting that the numbers in-
dicate the execution order.
in Section 4.1). Every agent work like an expert sys-
tem, where the basic parameters used by the MR and
ANL agents rules are e-value and score. CR agent
decides the suggestion based on the best results given
by the MR agents to recommend the annotation. All
this procedure is done at the collaborative layer. Fig-
ure 4 shows the pipeline of BioAgents executing for a
particular program (BLAST) and data base (nr) with
a gene identified.
5 EXPERIMENTS
In order to validate BioAgents new version, we used
data from three genome sequencing projects devel-
oped at the MidWest Region of Brazil: Functional and
Differential Genome from the Paracoccidioides bra-
siliensis (Pb) fungus (https://dna.biomol.unb.br/Pb-
eng/), Genome Project of Paullinia cupana plant
(guaran´a) (https://dna.biomol.unb.br/GR/) and
Genome Project of the Anaplasma marginale rick-
ettsia (https://www.biomol.unb.br/anaplasma/servlet/
IndexServlet).
Considering the Genome Project Pb, the analyzed
data were extracted from BLAST executed with nr,
COG and GO data bases; and from BLAT with S.
cereviseae and S. pombee fungi data bases. For the
Genome Project Guaran´a, we used BLAST executed
with nr, KOG and SwissProt data bases. We have used
the same nr data base adopted for the annotation on
both projects, Pb and Guaran´a, in order to compare
with BioAgents suggestions.
Since the Genome Project Anaplasma was not
manually annotated, we used BioAgents to support
the annotation task. For this project we used BLAST
with nr and Anaplasma marginalis St. Maries data
base. For the three projects, PORTRAIT was used if
BLAST and BLAT did not find any similar sequences
as presented in Section 4.1.
From the Genome Project Pb, 6, 107 sequences
were analyzed (Table 1). From these, 2, 820 genes
were manually annotated by the biologists, and 3, 287
were not. Note that 3,040 annotations were sug-
gested by BioAgents, being 1,746 correct when com-
pared to the 2,820 manual annotations of the Genome
Project Pb, which corresponds to 57.48% of correct
suggestions. Observe that for the 3,287 not manu-
ally annotated genes, 533 were suggested by BioA-
gents. From the 3,067 not identified as putative pro-
teins, 447 were identified as ncRNA. According to
the biologists, these are good results that can be even
improved as the agent knowledge bases are refined.
Also, we consider the correctness method used very
naive (three equal strings), which demands semantic
improvement.
Table 1: Results of BioAgents applied to the Genome
Project Pb.
Number of genes 6,107
Number of genes manually annotated 2,820
Number of annotations suggested by
BioAgents 3,040
Number of annotations correctly 1,746/
indicated by BioAgents 3,040
Percentage of correct suggestions
(related to the manual annotations) 57.48%
Number of annotations suggested for 533/
genes not manually annotated 3,287
Number of sequences not identified as
putative proteins 3,067
Number of ncRNAs 447
Percentage of ncRNAs 14.57%
We analyzed 8,597 sequences of the Genome
Project Guaran´a (Table 2). From these, 7,725 genes
were manually annotated by the biologists and 872
were not. Note that 6,354 annotations were suggested
A MULTI-AGENT TOOL TO ANNOTATE BIOLOGICAL SEQUENCES
229
by BioAgents, being 3,626 correct when compared to
the 7,725 manual annotations, which corresponds to
57.07% of correct suggestions. From the 2, 243 not
identified as putative proteins, 1,217 were identified
as ncRNAs. The ncRNAs results of 54.25% proved
the importance to use a ncRNA algorithm like POR-
TRAIT in the Guaran´a Project.
Table 2: Results of BioAgents applied on the Genome
Project Guaran´a.
Number of genes 8,597
Number of genes manually annotated 7,725
Number of annotations suggested by
BioAgents 6,354
Number of annotations correctly 3, 626/
indicated by BioAgents 6,354
Percentage of correct suggestions
(related to the manual annotations) 57.07%
Number of annotations suggested for 367/
genes not manually annotated 872
Number of sequences not identified as
putative proteins 2,243
Number of ncRNAs 1,217
Percentage of ncRNAs 54.25%
For the Genome Project Anaplasma, BioAgents
suggested 2,401 annotations for a total of 3,214
ORFs (Table 3), corresponding to 74.70% of sugges-
tions. This was an expected result since one of the
used data base was from the same already annotated
organism Anaplasma marginalis St. Maries. From
the 813 not identified as putative proteins, 502 were
identified as ncRNAs, which corresponds to 61.74%.
Table 3: Results of BioAgents applied on the Genome
Anaplasma Project.
Number of contigs 773
Number of ORFs at the contigs 1,541
Number of annotations to ORFs
(contigs) suggested by BioAgents 1,343
Number of singlets 1,041
Number of ORFs on singlets 1,673
Number of annotations to ORFs
(singlets) suggested by BioAgents 1,058
Number of ORFs 3,214
Number of ORF annotations
suggested by BioAgents 2,401
Percentage of suggestions 74.70%
Number of not identified as putative proteins 813
Number of ncRNAs 502
Percentage of ncRNAs 61.74%
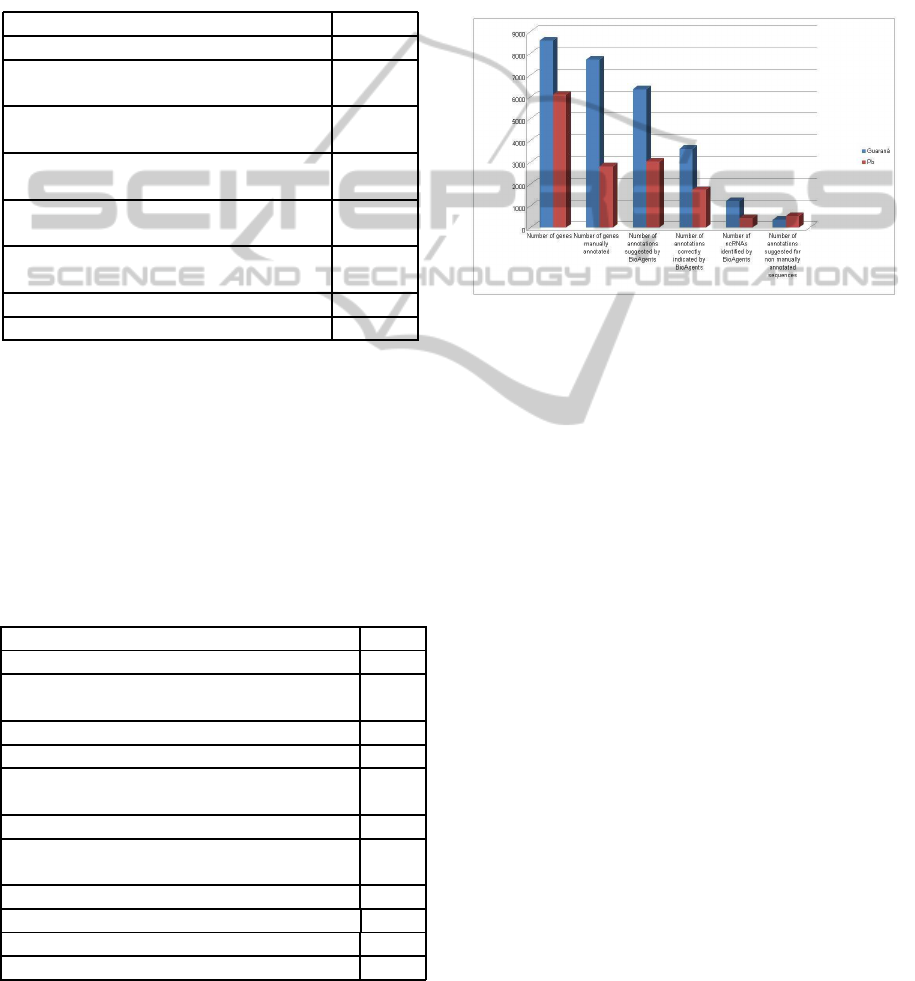
Figure 5 shows the results of Genome Project Pb
and Genome Project Guaran´a according to Tables 1
and 2. Note that the adopted rules use only the e-value
and score computed by BLAST and BLAT. Particu-
larly, we do not consider a minimum percentage of the
score, since this information was not used by the biol-
ogists in their manual annotation. Programs to iden-
tify ncRNAs were not used in the genome projects
used in our experiments, although BioAgents have
used PORTRAIT. In addition, the adopted basic rules
lead to good results.
Figure 5: Comparisons among the results of Genome
Project Pb and Genome Project Guaran´a.
6 CONCLUSIONS AND FUTURE
WORK
In this article, we presented a new version of a so-
phisticated prototype called BioAgents, a MAS to
annotate biological sequences in genome projects.
The annotation process is based on heterogeneous
and dynamic environment, and biologists can ana-
lyze sequences of interest in order to confirm com-
putational results. BioAgents uses different and dis-
tributed databases, with data being continuously mod-
ified, which fits well to the multi-agent approach.
BioAgents has agents specialized on distinct tasks, so
that they can act independently,using their knowledge
represented through specific inference rules.
As mentioned before, considering the new sce-
nario of massively parallel automatic sequencers,
with billions of little fragments, BioAgents can
strongly help to improve the quality of the annota-
tion process. As far as we know, the majority of the
systems developed to support annotation like the ones
cited in Section 3 are organisms specific (virus or pro-
teins) and do not have a reasoning mechanism to sug-
gest annotation. In addition, they do consider ncR-
NAs during the annotation phase.
BioAgents can be improved in many different
ways. Including other algorithms that analyse dif-
ICAART 2011 - 3rd International Conference on Agents and Artificial Intelligence
230

ferent characteristics of the sequences, as FASTA for
example. The improvement of agents knowledge is
necessary to achieve a higher accuracy for the sug-
gestions proposed by BioAgents. Also more complex
semantic and ontological methods would improve the
suggestions in BioAgents. Finally, BioAgents can
be configurable to a more distributed implementation
system using clusters, grids or cloud computing re-
sources. In the experimental aspects, we plan to use
BioAgents in high throuput genome projects, that are
beginning in the MidWest Region of Brazil.
REFERENCES
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and
Lipman, D. J. (1990). Basic local alignment search
tool. J Mol Biol, 215(3):403–410.
Arrial, R. T., Togawa, R. C., and Brigido, M. M. (2007).
Outlining a Strategy for Screening Non-coding RNAs
on a Transcriptome Through Support Vector Ma-
chines. In Sagot, M.-F. and Walter, M. E. T., editors,
BSB, volume 4643 of Lecture Notes in Computer Sci-
ence, pages 149–152. Springer.
Bellifemine, F. L., Caire, G., and Greenwood, D. (2007).
Developing Multi-Agent Systems with JADE. John Wi-
ley & Sons Ltd. ISBN 978-0-470-05747-6.
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell,
J., and Wheeler, D.L. (2008). Genbank. Nucleic Acids
Res, 36(Database issue).
do Nascimento, L. V. and Bazzan, A. L. C. (2004). An
agent-based system for re-annotation of genomes.
In III Brazilian Workshop on Bioinformatics (WOB),
pages 41–48.
Eddy, S. R. and Durbin, R. (1994). Rna sequence anal-
ysis using covariance models. Nucl. Acids Res.,
22(11):2079–2088.
Forgy, C. L. and Shepard, S. J. (1987). Rete: a fast match
algorithm. AI Expert, 2(1):34–40.
Hill, E. F. (2003). Jess in Action: Java Rule-Based Systems.
Manning Publications Co., Greenwich, CT, USA.
Iacono, M., Villa, L., Fortini, D., Bordoni, R., Im-
peri, F., Bonnal, R. J. P., Sicheritz-Ponten, T.,
Bellis, G. D., Visca, P., Cassone, A., and Carat-
toli, A. (2008). Whole-genome pyrosequencing of
an epidemic multidrug-resistant acinetobacter bau-
mannii strain belonging to the european clone ii
group. Antimicrobial Agents and Chemotherapy,
52(7). doi:10.1128/AAC.01643-07.
Kent, W. J. (2002). Blat–the blast-like alignment tool.
Genome research, 12(4):656–664.
Lima, R. S. (2007). Multiagent System for Manual An-
notation in Genome Sequencing Projects. Master’s
thesis, Department of Computer Science, University
of Bras´ılia. Available in Portuguese at http://mono-
grafias.cic.unb.br/dspace/handle/123456789/111.
Mardis, E. R. (2008). Next-generation dna sequencing
methods. Annual Review of Genomics and Human
Genetics, 9(1):387–402.
Moore1, M. J., Dhingra, A., Soltis, P. S., Shaw, R.,
Farmerie, W. G., Folta, K. M., and Soltis, D. E.
(2006). Rapid and accurate pyrosequencing of an-
giosperm plastid genomes. BMC Plant Biology, 6(17).
doi:10.1186/1471-2229-6-17.
Pearson, W. R. and Lipman, D. J. (1988). Improved tools for
biological sequence comparison. Proceedings of the
National Academy of Sciences of the USA, 85:2444–
2448.
Pop, M. and Salzberg, S. L. (2008). Bioinformatics chal-
lenges of new sequencing technology. Trends in ge-
netics : TIG, 24(3):142–149.
Ralha, C. G. (2009). Towards the integration of multia-
gent applications and data mining. In Cao, L., editor,
Data Mining and Multi-agent Integration, pages 37–
46. Springer Science + Business Media. ISBN: 978-
1-4419-0521-5, DOI: 10.1007/978-1-4419-0522-2 2.
Ralha, C. G., Schneider, H. W., Fonseca, L. O., Walter,
M. E., and Br´ıgido, M. M. (2008). Using BioAgents
for Supporting Manual Annotation on Genome Se-
quencing Projects. In BSB’08: Proceedings of the
3rd Brazilian symposium on Bioinformatics-Lecture
Notes in Bioinformatics, volume 5676, pages 127–
139, Berlin, Heidelberg. Springer-Verlag.
Ralha, C. G., Schneider, H. W., Walter, M. E. M. T.,
and Bazzan, A. L. C. (2010). Reinforcement
learning method for bioagents. In XI Brazilian
Symposium on Artificial Neural Network, SBRN
2010, So Bernardo do Campo, So Paulo, October
23-28, pages 1–6. IEEE Computer Society Press.
http://www.jointconference.fei.edu.br/index.htm.
Schroeder, L. F. and Bazzan, A. L. C. (2002). A multi-
agent system to facilitate knowledge discovery: an
application to bioinformatics. In Proceedings of the
Workshop on Bioinformatics and Multi-Agent Systems
(BIXMAS’2002), pages 44–50, Bologna, Italy.
Solda, G., Makunin, I. V., Sezerman, O. U., Corradin, A.,
Corti, G., and Guffanti, A. (2009). An ariadne’s thread
to the identification and annotation of noncoding rnas
in eukaryotes. Brief Bioinform, 10(5):475–489.
Wang, Z., Gerstein, M., and Snyder, M. (2009). Rna-seq: a
revolutionary tool for transcriptomics. Nature Reviews
Genetics, 10(1):57–63.
Weiss, G. (2000). Multiagent Systems: A Modern Approach
to Distributed Artificial Intelligence. The MIT Press.
ISBN 0-262-73131-2.
Wooldridge, M. (2009). Introduction to MultiAgent Sys-
tems. John Wiley & Sons, 2nd edition. ISBN 978-0-
470-51946-2.
A MULTI-AGENT TOOL TO ANNOTATE BIOLOGICAL SEQUENCES
231
