
3D Data Hiding for Enhancement and Indexation on
Multimedia Medical Data
N. Tournier
1,2
, G. Subsol
1
, W. Puech
1
and J-P. Pedeboy
2
1
Universit
´
e de Montpellier 2, LIRMM, CNRS
161 rue Ada, Montpellier, France
2
STRATEGIES S.A., 41-43 rue de Villeneuve
Parc des Affaires SILIC - BP 80429, Rungis, France
Abstract. In medical applications, large quantities of multimedia data are ex-
changed such as 3D data acquired either by volume (CT-scan) or surface (laser
range) scanning. The increasing of the numerical data using raises some unsolved
issues. As for us, we are interested in the protection and the enhancement of mul-
timedia content by insertion of hidden message. According to Koller et al. [7]
some of these challenges are:
– Metadata embedding;
– Indexing and searching in database.
Data hiding may be a solution for these main applications in the medical domain.
It is possible to embed metadata, with security for confidential data or for index-
ing area in a media, without increasing the size of the file.
1 Introduction
The main interest of data hiding is to embed additional information without increasing
the file size and keeping the compatibility with the norms and the standards such as
JPEG for image, DICOM for medical image, MPEG for video, etc.
In this Section, we propose to introduce briefly the data hiding principle and we
present some medical application.
1.1 Data Hiding Principle
The principle is to embed a message into the useful data of the multimedia file and to
be able to extract or recognize it from the watermarked file. We can divide the insertion
process in two parts: the synchronization and the embedding.
Synchronization allows us to know where the encoded message can be embedded
in the host signal. It can be considered as a preparation of the media to the embed-
ding. The host signal can be represented in other domain, such as a frequential domain
(Discrete Cosines Transform, Fourier Transform), a wavelet domain (Discrete Wavelet
Transform) as a function of the application and the chosen method. The aim is to be
able to recognize the same subspace, called insertion subspace, before and after the
watermarking.
Tournier N., Subsol G., Puech W. and Pedeboy J..
3D Data Hiding for Enhancement and Indexation on Multimedia Medical Data.
DOI: 10.5220/0003311800430051
In Proceedings of the 2nd International Workshop on Medical Image Analysis and Description for Diagnosis Systems (MIAD-2011), pages 43-51
ISBN: 978-989-8425-38-6
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
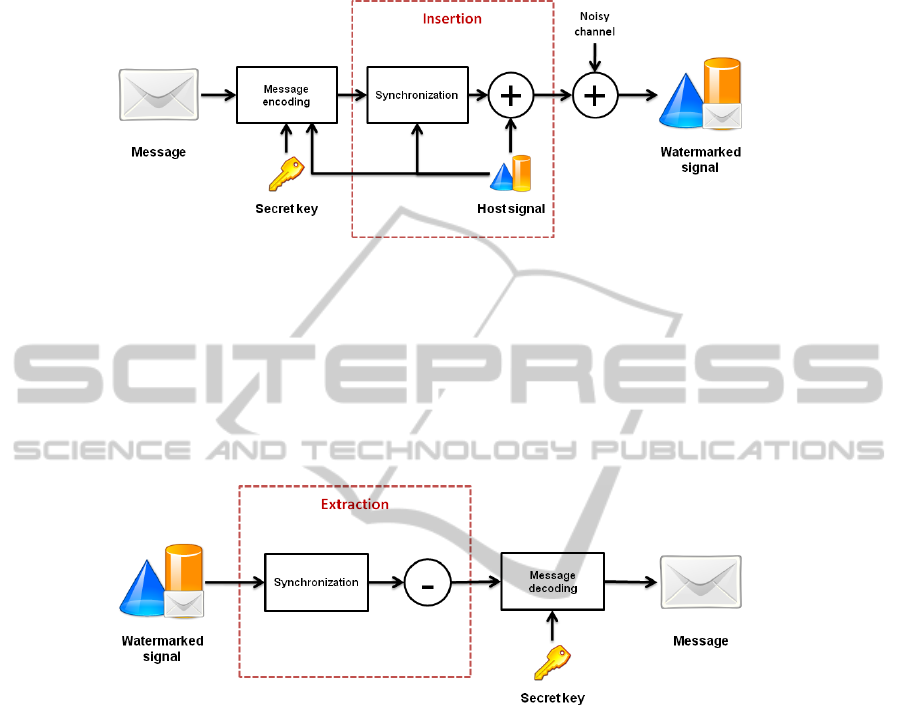
Fig. 1. Watermarking Scheme.
The message is encoded as a function of a secret key and the host signal. The em-
bedding consists in merging the encoded message and the host signal in the subspace
domain. The embedding principle is illustrated on Fig. 1.
On the watermarked data, we need to extract the embedded message. For the extrac-
tion process, first we synchronize the watermarked data to find the insertion subspace
again and we extract the message depending on the insertion method. The extraction
principle is illustrated on Fig. 2.
Fig. 2. Extraction Scheme.
1.2 Data Hiding Applications in Healthcare
The applications are various and depend on compromises between the capacity of the
message to embed, the insertion robustness, the modification imperceptibility [11], the
algorithm security [6] and the complexity. As the data are voluminous, the algorithms
must be fast with a low complexity.
One of the main application of data hiding is communication, a transmission of a
secret message. It looks like cryptography approaches. In data hiding we do not want to
secure the media to transmit, the media is a carrier for the message we want to send. In
this condition, the watermark algorithm must be robust and secured. For example, in a
medical context, it could be interesting to embed the identity of the patient into the 3D
image acquired by a CT-scan. For privacy, we need to keep anonymous its identity for
a secured transmission of the data. We can note cryptography and data hiding can be
combined for these applications [12].
44

Data hiding can be used for enhance the media with metadata. It is quite easy to add
metadata in a file, but we do not want to increase the size of the media because more
the data are voluminous; more it is difficult to store them. Such as the embedding of
indexing information [3] in order to identify the media. It can be also possible to carry a
diagnostic resume, a description of the image, some quotes or remarks in order to have
all the main information available on the same file. But there are some constraints for
example in ROI medical image, where the image must not be disrupt in the ROI areas
[14].
For further information on data hiding application in healthcare, [5] made a review
of image watermarking applications.
Data hiding is useful in medical application. Nowadays, there are a lot of well-
known techniques in 2D watermarking. In the 3D domain, the topic is more difficult
because the synchronization issue is more difficult. There is not any way to scan the 3D
object obviously rather than in 1D scanning the audio with a pseudo-random clock; or
in 2D scanning the image line by line for example.
In the following Section, we propose data hiding system for 3D objects. First, we
introduce a watermark technique on 3D images acquired by CT-scan in Section 2 and in
Section 3 we deal with a watermark technique applied on 3D meshes. The presentation
of the techniques is validated by experimental results in the respective sections.
2 3D Image Data Hiding
2.1 2D Data Hiding on CT-scan before 3D Reconstruction
For this approach, the idea is to use well-known 2D watermarking technique for 3D
object. First, the object is acquired by volume (CT-scan) in order to have a 3D image.
Each slice is equivalent to a 2D image that we can watermark by classical techniques.
We illustrate on Fig. 3 some slices of a 3D image acquired by CT-scan.
From the 3D image we are able to build a 3D mesh by computing the iso-surface.
We illustrate the reconstruction of the skin (Fig. 4.a.) and the skeleton (Fig. 4.b.) from
our 3D image. The aim is to watermark the 3D image such as there is no perceptual
difference on the 3D object.
In our condition, we have 3D images
3
composed by 249 slices (512 × 512 pixels).
We can watermark each slice with the same message, or independently with a different
message for each one. It depends on the final application. For example, if we prefer to
have a robust watermarking scheme, we will watermark the same message in each slice;
else if we want to enhance the media with metadata, we prefer to have an high capacity
watermarking scheme so we embed different message in each slice.
In that case, we just want to prove that the watermark keep a good quality on the 3D
reconstruction. We choose a well-known technique, by substitution of the LSB value of
the pixels [2].
When the images are watermarked, we reconstruct the object from the 3D image
into a mesh [8].
3
http://pubimage.hcuge.ch:8080/.
45
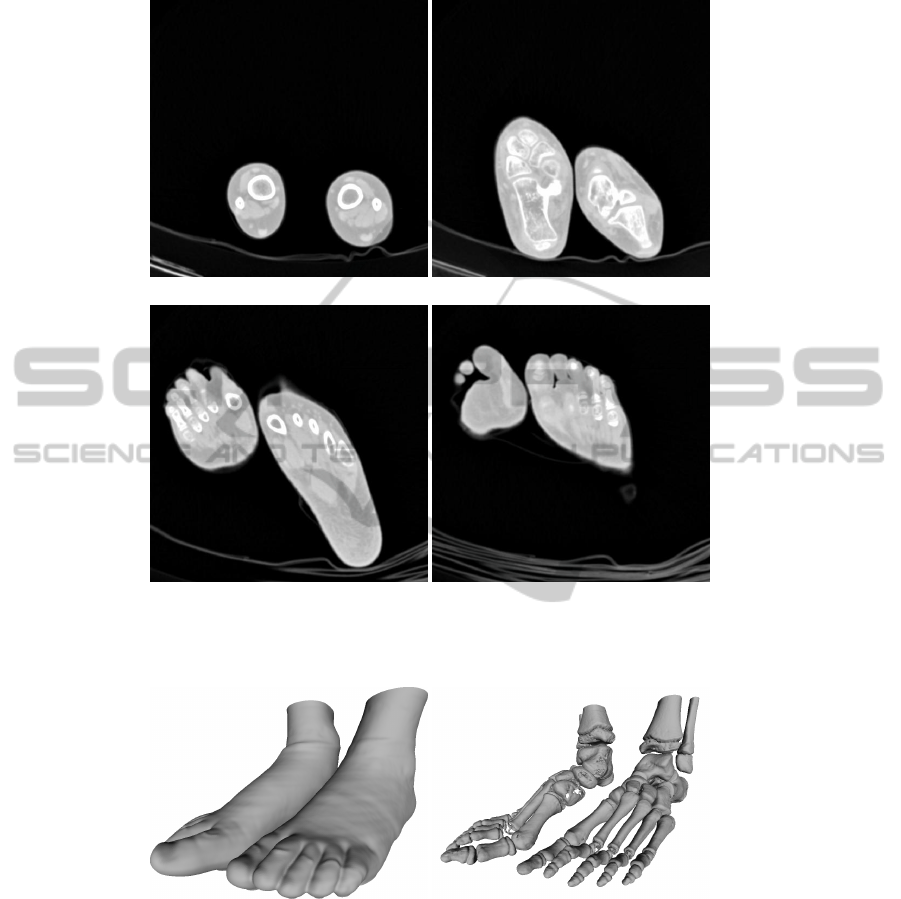
(a) (b)
(c) (d)
Fig. 3. An example of some 3D image slices: a) A represention of the slice #1, b. the slice #130,
c. the slice #200 and d. the slice #230.
(a) (b)
Fig. 4. An example of 3D mesh reconstruction from the 3D image: a) The skin reconstruction
composed by 663
0
072 vertices and 1
0
126
0
466 faces; b) The skeleton one composed by 804
0
073
vertices and 1
0
577
0
853 faces.
For the extraction procedure, we just need to extract the message from the 3D image
by reading the LSB value of each watermarked pixel. It is a fragile technique with high
capacity, fast (low complexity) and quite efficient to enhance 3D images with metadata.
46
2.2 Experimental Results
From the 3D image composed by 249 slices (512 × 512 pixels) we embed in each slice
30ko. With the LSB watermarking method, associated with a pseudo-random image
scanning; we can embed one bit per pixel. In these conditions, the capacity is fixed at
0.9265 bit per pixel.
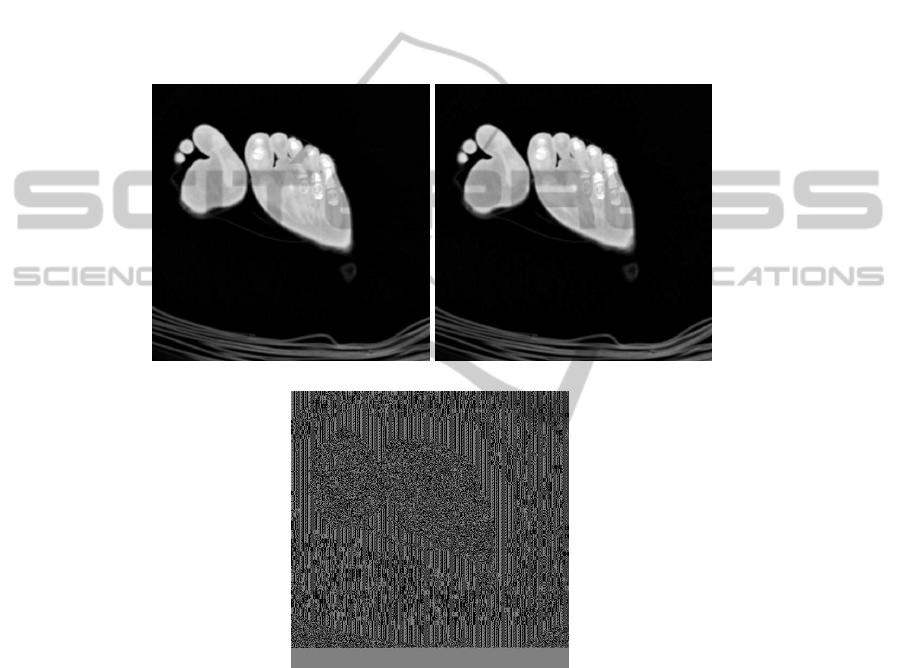
First, we are looking for the quality of the watermark on each image. We compute
the PSNR (Peak Signal Noise Rate) between the original slice and the watermark one.
The results are very good, the PSNR equals to 51.63 dB in average for an high ca-
pacity application. There is no perceptual difference between a slice and the respective
watermarked one (Fig. 5).
(a) (b)
(c)
Fig. 5. Comparison between: a) The original slice #230; b) The watermarked image; c) The
difference image.
The main aim is to know what is the impact of the watermark on the 3D mesh
reconstruction. We build the 3D mesh, by computing iso-surface on the watermarked
3D image and we compare the quality of the mesh obtained by the original image and
this one.
To quantify the quality we use the Hausdorff distance, that computes the longest dis-
tance of the shortest distance between a vertex in the first mesh to a vertex in the second
47
one. For the skeleton mesh and for the skin mesh, the Hausdorff distance equals to 0.00.
That means there is no difference between these two meshes and we are able to extract
the watermark from the 3D image. It is a very high capacity technique, nevertheless we
can not extract the message from the 3D mesh.
3 3D Mesh Data Hiding
3.1 3D Data Hiding using Euclidean Minimum Spanning Tree
In Section 2, we have presented a 2D watermark technique used for 3D objects. We
remark the extraction process must be done from the 3D image and not from the 3D
mesh. In this Section, we want to watermark the mesh. Various techniques exist, most
of them are robust to geometrical rigid transformations. In this Section, we present an
original 3D watermarking approach that does not move any vertex [1].
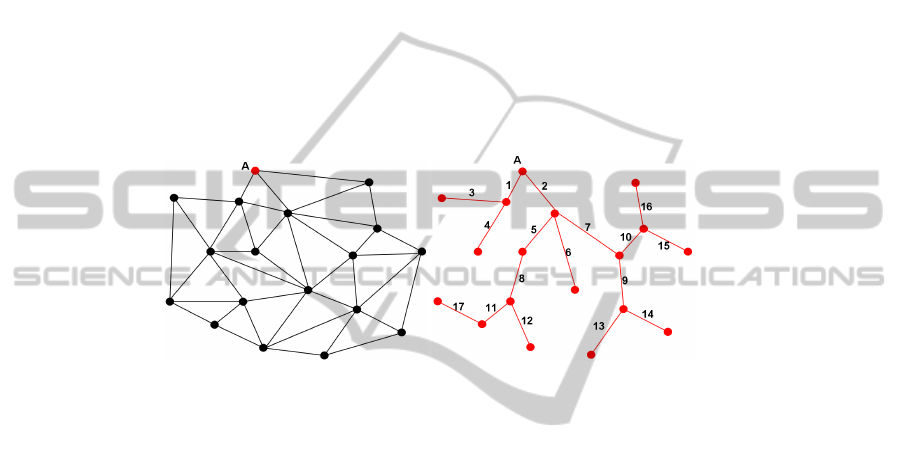
(a) (b)
Fig. 6. An example of a mesh: a) The EMST of the mesh; b) We can remark connections in the
EMST are not necessarily edges in the mesh.
Few 3D watermarking approaches do not move any vertex [1], [10], [9]. The origi-
nality of [1] is to watermark a message by editing the connections in the mesh without
increasing the file size and without moving the vertices. In order to synchronize the
message they compute an Euclidean minimum spanning tree (EMST) illustrated Fig. 6.
The EMST is unique, and from a seed vertex v
0
, the path of the vertices is also unique.
This can be an interesting synchronization tool because we can cover the mesh with a
unique path.
In the EMST, they are searching quadruples, a father-vertex and three sons, to em-
bed one bit. The insertion is quite simple, if we want to embed a 0-bit the edge of the
EMST is chosen. Otherwise, if we want to embed a 1-bit, the other edge is in the EMST.
The insertion process is illustrated Fig. 7.
In order to not create important visual distortions and synchronizations issues, the
quadruples must verify the following conditions:
– Coplanarity: the measure of the angle formed by the two triangles (Fig. 8);
– Convexity: the quadruple must be convex in order to cover quite the same surface
(Fig. 9);
– Covering: if two quadruples are neighbours only one of them is used for the em-
bedding.
48
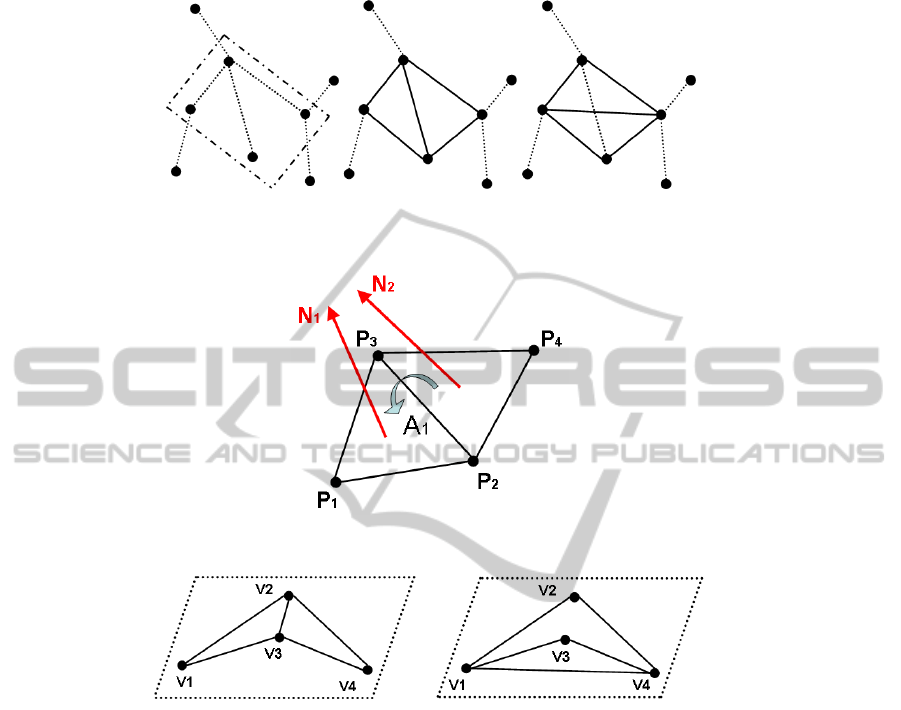
(a) (b) (c)
Fig. 7. Illustration of the EMST (a), the mesh when a 0-bit is embedded (b) and when a 1-bit (c).
Fig. 8. Representation of the coplanarity condition.
(a) (b)
Fig. 9. Representation of the watermarking impact on a quadruple not convex before the water-
mark (a) and after (b). The covered area is different and there is a topological problem: v
3
must
not be inside of the triangle (v
1
, v
2
, v
4
).
3.2 Experimental Results
We present some results on our 3D Skeleton. We illustrate, Fig. 10.a the simplified
3D mesh and its EMST Fig. 10.b. During the watermarking process, the vertices are
not moving, the cloud is the same, only the connectivity is changed. Indeed, there is
not any distortion due to the vertex displacement. So we compute the Hausdorff error,
including the connectivity distortion. The Hausdorff distance is computed on the center
of the triangles and not on the vertices and normalised as a function of the bounding
box diagonal computed with METRO [4].
As a function of the coplanarity threshold, we can embed more information in the
mesh. More the threshold is low, more the connectivity distortion is better but the capac-
49

(a) (b)
Fig. 10. An example of EMST applied on a simplified 3D mesh Skeleton (48
0
711 vertices and
98
0
614 faces).
ity maximum decrease because we select fewer quadruples. For a coplanarity threshold
fixed at 10
◦
we can embed a message of 575 bits maximum in Skeleton and for 5
◦
the
maximum capacity is up to 264 bits.
In order to compare the distortion in the mesh, we fix the capacity of insertion to
4 · 10−3 bits per vertex (i.e. 216 bits on Skeleton). After the embedding we compute
the Hausdorff distortion. For a coplanarity thresholds equals to 10
◦
we get 3.94 · 10
−4
and 5
◦
we have 1.08 · 10
−4
. That means for a diagonal of 20 cm, the maximum error is
0.08 mm.
4 Conclusions
We have presented a watermarking overview for medical application, in order to protect
the 3D images or to enhance the 3D object by embedding metadata. In medical appli-
cation the data are large for each patient, so we need to develop fast algorithms without
increasing the size of the data.
We have introduced 2D watermarking techniques used for 3D images. It is a low
complexity algorithm with high complexity to enhance the 3D model. But it is very
fragile to noise addition and it is impossible to extract the message from the 3D mesh.
Then, we have presented a 3D watermark method based on a mesh. The originality
is to watermark a message without moving any vertices and without increasing the file
size. The synchronization is made by the localization of quadruples in the EMST of the
vertex cloud. The method is also fragile with low capacity of insertion, but the 3D mesh
is watermarked. Nevertheless, the complexity is very high (quadratic as a function of
the number of vertices).
It is very fragile to noise addition, so we focus on this method to improve it by
studying the possible displacement of the vertex in order to be more robust. As a func-
tion of the Prim’s algorithm and the seed vertex, we are able to know how can a vertex
be moved without editing the connections in the EMST [13]. Indeed, we watermark our
message in the most robust areas.
We study the EMST structure in order to find new robust criterion in order to select
robust areas and improve the computational time.
50

References
1. P. Amat, W. Puech, S. Druon, and J.P. Pedeboy. Lossless 3D Steganography Based on MST
and Connectivity Modification. Signal processing. Image communication, 25(6):400–412,
2010.
2. C.K. Chan and LM Cheng. Hiding Data in Images by Simple LSB Substitution. Pattern
Recognition, 37(3):469–474, 2004.
3. S. Cheng, Q. Wu, and K.R. Castleman. Non-Ubiquitous Digital Watermarking for Record
Indexing and Integrity Protection of Medical Images. In Image Processing, 2005. ICIP 2005.
IEEE International Conference on, volume 2. IEEE, 2005.
4. P. Cignoni, C. Rocchini, and R. Scopigno. Metro: measuring error on simplified surfaces. In
Computer Graphics Forum, volume 17, pages 167–174. Wiley Online Library, 1998.
5. G. Coatrieux, L. Lecornu, B. Sankur, and C. Roux. A Review of Image Watermarking Ap-
plications in Healthcare. In Proc. of the 28th International Conference IEEE Engineering in
Medicine and Biology Society, 2006.
6. A. Kerckhoffs. La Cryptographie Militaire. Journal des sciences militaires, 9(5-38):161–191,
1883.
7. D. Koller, B. Frischer, and G. Humphreys. Research Challenges for Digital Archives of 3D
Cultural Heritage Models. Journal on Computing and Cultural Heritage (JOCCH), 2(3):1–
17, 2009.
8. W. E. Lorensen and H. E. Cline. Marching Cubes: A High Resolution 3D Surface Construc-
tion Algorithm. In Proceedings of the 14th annual conference on Computer graphics and
interactive techniques, page 169. ACM, 1987.
9. X. Mao, M. Shiba, and A. Imamiya. Watermarking 3D Geometric Models Through Triangle
Subdivision. In Proc. SPIE, volume 4314, pages 253–260, 2001.
10. R. Ohbuchi, H. Masuda, and M. Aono. Watermaking Three-Dimensional Polygonal Models.
In Proceedings of the Fifth ACM International Conference on Multimedia, pages 261–272.
ACM New York, NY, USA, 1997.
11. B. Planitz and A. Maeder. Medical Image Watermarking: A Study on Image Degradation.
In Proceedings of the Workshop on Digital Image Computing: Technqiues and Applications,
pages 3–8, 2005.
12. W. Puech and J. M. Rodrigues. A New Crypto-Watermarking Method for Medical Images
Safe Transfer. In Proc. of the 12th European Signal Processing Conference, pages 1481–
1484, 2004.
13. N. Tournier, W. Puech, G. Subsol, and J-P. Pedeboy. Sensitivity Analysis of Euclidean Min-
imum Spanning Tree. In SPIE’10: 3D Image Processing (3DIP) and Applications, San Jose,
USA (CA), 2010.
14. A. Wakatani. Digital Watermarking for ROI Medical Images by Using Compressed Sig-
nature Image. In System Sciences, 2002. HICSS. Proceedings of the 35th Annual Hawaii
International Conference on, pages 2043–2048. IEEE, 2002.
51
