
EVOLUTIONARY SUPPORT VECTOR MACHINE
FOR PARAMETERS OPTIMIZATION APPLIED
TO MEDICAL DIAGNOSTIC
Ahmed Kharrat, Nacéra Benamrane
University of Sfax, National Engineering School, Computer & Embedded Systems Laboratory (CES)
B.P 1173, Sfax 3038, Tunisia
Department of Computer Science, Faculty of Science, Vision and Medical Imagery Laboratory U.S.T.O.
B.P 1505, EL-Mnaouer, Oran, 31000, Algeria
Mohamed Ben Messaoud, Mohamed Abid
University of Sfax, National Engineering School, Laboratory of Electronics and Information Technologies
Computer & Embedded Systems Laboratory (CES), B.P 1173, Sfax 3038, Tunisia
Keywords: Support vector machine, Classification, Genetic algorithm, Parameters optimisation, Feature selection.
Abstract: The parameter selection is very important for successful modelling of input–output relationship in a
function classification model. In this study, support vector machine (SVM) has been used as a function
classification tool for accurate segregation and genetic algorithm (GA) has been utilised for optimisation of
the parameters of the SVM model. Having as input only five selected features, parameters optimisation for
SVM is applied. The five selected features are mean of contrast, mean of homogeneity, mean of sum
average, mean of sum variance and range of autocorrelation. The performance of the proposed model has
been compared with a statistical approach. Despite the fact that Grid algorithm has fewer processing time, it
does not seem to be efficient. Testing results show that the proposed GA–SVM model outperforms the
statistical approach in terms of accuracy and computational efficiency.
1 INTRODUCTION
Selecting vital features out of the original feature set
constitutes a challenging task. Supervised
classification models are usually used together with
optimization algorithms for feature selection, in
which classification accuracies are used as fitness
evaluation of the selected feature subsets (Kharrat,
2010a), (Kharrat, 2010b). In this work, we develop
an approach to optimize the support vector machine
parameters which combines the merits of support
vector machine (SVM) and genetic algorithm (GA).
Later we compare our proposed method with the
statistical approach to show its superiority in terms
of computational efficiency. Among the statistical
methods, we use the Grid algorithm.
Support vector machine (Boser, 1992) is among
the most popular classifiers used in supervised
learning. Its principle lies in constructing an optimal
separating hyperplane to maximize the margin
between two classes of data. The choice of margin
cost C and kernel parameters have an important
effect on the performance of SVM classifier
(Chapelle, 2003). The optimal parameters that lead
to the minimal generalization error are data-
dependent. Two-dimensional grid is usually used to
tune a pair of parameters such as C and
γ
(Gaussian
function width in RBF kernel) due to its complexity.
Even after parameter tuning, SVM classifier might
deliver poor accuracy in classifying some particular
datasets. Having as input these five selected features,
we build a technique to optimize SVM parameters.
There are many optimization techniques that have
been used. One of the favored choices is Genetic
Algorithm (GA).
Our present work depicts GA as an efficient tool
only to optimize the SVM parameters, without
degrading the SVM classification accuracy. The
201
Kharrat A., Benamrane N., Ben Messaoud M. and Abid M..
EVOLUTIONARY SUPPORT VECTOR MACHINE FOR PARAMETERS OPTIMIZATION APPLIED TO MEDICAL DIAGNOSTIC.
DOI: 10.5220/0003326902010204
In Proceedings of the International Conference on Computer Vision Theory and Applications (VISAPP-2011), pages 201-204
ISBN: 978-989-8425-47-8
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

proposed method performs parameters optimization
setting in an evolutionary way. In the literature,
some GA-based feature selection methods were
proposed (Raymer, 2000), (Yang and Honavar,
1998), (Salcedo-Sanz, 2002). However, these papers
focused on feature selection and did not deal with
parameters optimization for the SVM classifier.
Frohlich et al. (Frohlich and Chapelle, 2003)
proposed a GA-based feature selection approach that
used the theoretical bounds on the generalization
error for SVMs. In Frohlich’s paper, the SVM
regularization parameter can also be optimized using
GAs.
The remainder of this paper is organized as follows.
Section 2 describes parameters optimization
concepts. Section 3 describes system architecture for
the proposed GA-SVM model. Section 4 presents
the experimental results from using the proposed
method to classify real world dataset and to depict
the results. Section 5 summarizes and draws a
general conclusion.
2 PARAMETERS OPTIMIZATION
APPROACHES
Many kernel functions are used to help the SVM in
obtaining the optimal solution. Among the used
kernel functions are the polynomial, sigmoid and
radial basis kernel function (RBF). Unlike a linear
kernel function, the RBF is generally applied most
frequently, because it can classify multi-dimensional
data. Our work applies an RBF kernel function in
the SVM to obtain optimal solution. Two major RBF
parameters applied in SVM, C and
γ
, must be set
appropriately. Parameter C represents the cost of the
penalty. Parameter
γ
has a much greater influence on
classification outcomes than C, because its value
affects the partitioning outcome in the feature space.
Values for parameters C and
γ
that lead to the
highest classification accuracy rate in this interval
can be found by setting appropriate values for the
upper and lower bounds (the search interval) and the
jumping interval in the search. As well as the two
parameters C and
γ
, other factors, such as the
quality of the feature's dataset, may influence the
classification accuracy rate. For instance, the
correlations between features influence the
classification result.
3 SYSTEM ARCHITECTURE
FOR THE PROPOSED
GA-SVM MODEL
Having the ability to perform better than other
kernels (Kharrat, 2010c), a radial basis function
(RBF) kernel SVM is adopted to establish support
vector classifiers. When using the RBF kernel, there
are two parameters (i.e. C and
γ
) to be tuned.
Improper selection of the two parameters is
demonstrated to cause over-fitting or under-fitting
problems. The proposed GA-based approach is
designed to optimize the parameters pair (C,
γ
) for
the SVM. To implement our proposed approach, this
research uses the RBF kernel function for the SVM
classifier because the RBF kernel function can
analysis higher-dimensional data. In our study,
classification accuracy, the numbers of selected
features are the criteria used to design a fitness
function. Thus, for the individual with high
classification, a small number of features produce a
high fitness value.
The procedure of the proposed GA-SVM method
is shown in “Figure 1”, and the steps are detailed as
follows.
Step1. Encode the parameters and the chromosomes
representing the SVM parameters as a binary string.
Step2. Initialize the population and produce the
initial population of chromosomes arbitrarily.
Step3. Include the five optimal features. Decode the
binary chromosomes into the corresponding
parameters representing the optimal features
(Kharrat, 2010a).
Step4. Find the selected parameters. Decode the
binary chromosomes into the corresponding
parameters representing the optimized pair (C
and
γ
).
Step5. Train the SVM model. Get the trained SVM
model after implementing the optimized parameters
to the training set. The test sets are guessed on the
basis of the trained SVM model.
Step6. Compute the fitness. For each chromosome,
the training set with corresponding feature subset
among the five, and parameters pair(C,
γ
) are
designed as entries to the SVM classifier to calculate
the k-fold cross-validation accuracy (fitness).
Step7. The termination criteria are that the fitness
value does not increase during the last M generations
or that the maximum generation number reaches N.
If the two criteria are achieved, then the iteration
process stops. Otherwise go to Step8.
VISAPP 2011 - International Conference on Computer Vision Theory and Applications
202
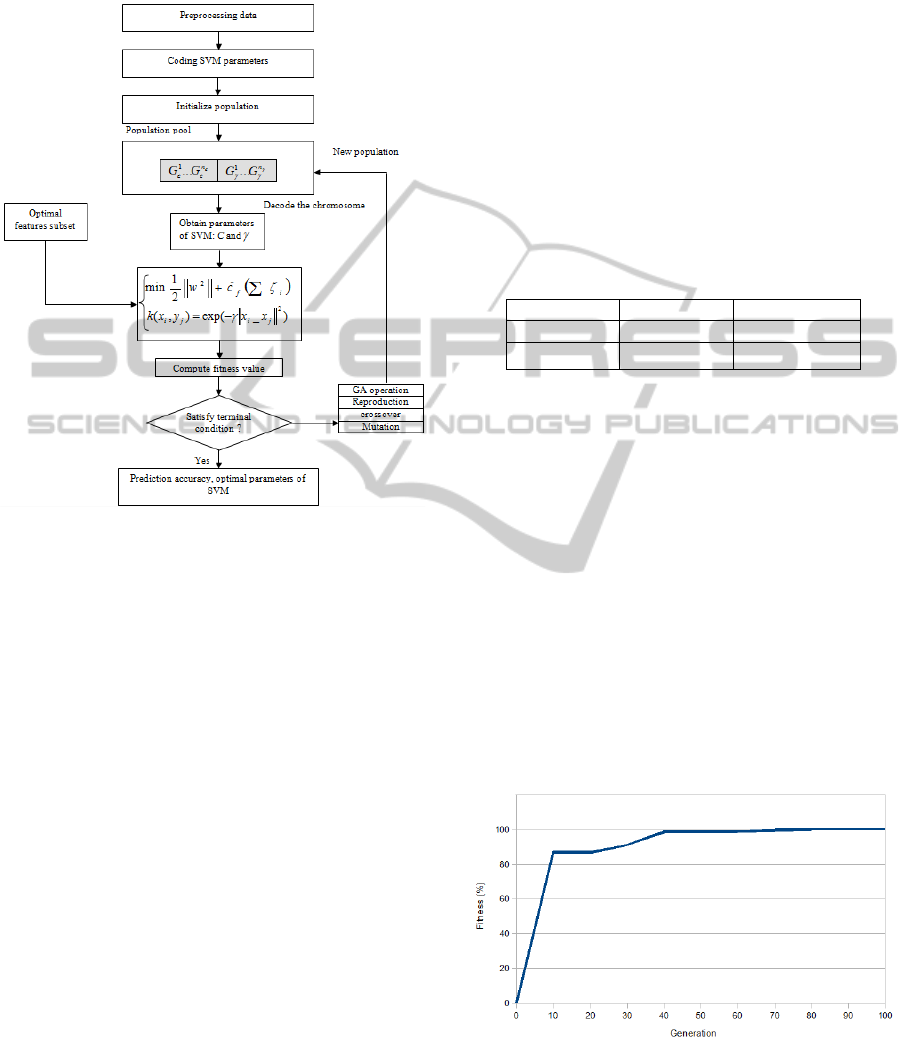
Step8. Perform genetic operations to generate an
offspring population. Genetic operations include:
crossover, mutation, and tournoi reproduction.
Figure 1: System architecture of the proposed GA-based
parameters optimization for support vector machine.
4 RESULTS
To evaluate the classification accuracy of the
proposed system in different classification tasks, we
tried a real human brain dataset from the Harvard
Medical School website (Keith and Alex, 1999).
These datasets consist in 83 images: 29 images are
normal and 54 belonging to pathological brain.
These normal and pathological benchmark
sagittal, axial, and coronal, images used for
classification are three weighted ones (enhanced T1,
proton density (PD) and T2) of 256×256 sizes and
acquired at several positions of the transaxial planes.
To guarantee valid results for making predictions
regarding new data, the dataset is further randomly
partitioned into training sets and independent test
sets via a k-fold cross validation. The advantages of
cross validation are that all of the test sets were
independent and the reliability of the results could
be improved. The data set is divided into k subsets
for cross validation. This study used k=5, meaning
that all of the data will be divided into 5 parts, each
of which will take turns at being the testing data set.
The other four data parts serve as the training data
set for adjusting the model prediction parameters.
The empirical evaluation was performed on Intel
core 2 duo machine, with 4GO RAM and a
processor speed of 2GHz, run under Windows XP
environment. To search for the best C and γ, the
Grid search algorithm is considered as a common
method. In the Grid algorithm, pairs of (C, γ) are
tried and the one with the best cross-validation
accuracy is chosen. The results from the proposed
method were compared with that from the Grid
algorithm. In all of the experiments we use 5-fold
cross validation to compute the accuracy of each
learned classifier.
Table 1: Range values for
γ
and C in GA and Grid
algorithm.
Parameter GA Grid algorithm
C ]0…2
20
] ]0…2
4
]
γ
[2
-20
…0[ ]0…2
2
]
The detail parameter setting for genetic algorithm is
as the following: population size 30, crossover rate
0.9, mutation rate 0.1, arithmetic crossover, tournoi
wheel selection, and elitism replacement. “Table 1”
states range values for both parameters
γ
and C in
genetic algorithm as well as grid algorithm.
“Figure 3” depicts a typical evolving process of the
GA. This process is characterized by three phases. In
the first phase the fitness value increases gradually
from the initialization value (86.9565%) to91.3043%
at the 30 generations. In the second phase the fitness
value rises more slowly: from 98.5491% at the 40
generation to 99.8897% at the 70 generation.
Whereas the third phase is characterized by stability.
When the generation number reaches 80, the
maximum fitness value is obtained and stays the
same (100%) until the 100 generation.
Figure 3: Iteration process of the GA for simultaneous
optimization (Generation/Fitness).
The final sentence of a caption must end with a
period.
EVOLUTIONARY SUPPORT VECTOR MACHINE FOR PARAMETERS OPTIMIZATION APPLIED TO MEDICAL
DIAGNOSTIC
203

Table 2: Genetically optimised SVM parameter for human brain dataset using GA-based approach and Grid algorithm.
Test period
GA-based approach Grid algorithm
C
γ
Accuracy Training days C
γ
Accuracy Training days
TP1 8 0.725 86.9565 % 10 0.5 0.0625 58.33 % 0.2
TP2 2383 0.0008 98.5682 % 8 7 2 97.19 % 0.8
TP3 19277 0.0087 99.8897 % 11 3 0.9 91.33 % 0.5
TP4 133455 0.2502 100 % 11 9 1.1245 95.33 % 0.6
TP5 131847 0.0009 100 % 11 8 2.019 94.71 % 0.9
The optimised values of SVM parameters for human
brain dataset using GA-based approach and Grid
algorithm corresponding to each TP are given in
“Table 2”. It can be observed that the optimum
values of these parameters vary significantly over a
wide range reflecting the superiority of GA to Grid
algorithm. In the GA, pairs of (C, γ) are tried and the
one with the best accuracy is chosen. To obtain the
best optimized pair of (C, γ), the process lasts
between 8 and 11 days, but the best accuracy is
achieved with a longer period. The parameters C
whose values exceed 2000 achieve high accuracy
surpassing 98% to 100%. In the Grid algorithm the
accuracy rate is low despite the short period of
training. Comparison of the obtained results of GA
with those of Grid algorithm demonstrates that GA-
SVM approach has a better classification accuracy
than the Grid algorithm tested.
5 CONCLUSIONS
This study presents an evolutionary computing
optimization approach, capable of searching for the
optimal parameter values for SVM by using a subset
of selected features. Compared with the statistical
approach, the proposed GA-based approach has
higher accuracy with fewer selected features. It
outperforms the statistical approach in terms of
computational efficiency. Moreover, the proposed
GA-based approach has proved to be effective in
optimizing parameters for the SVM. Results of this
study are obtained with an RBF kernel function.
However, other kernel parameters can also be
optimized using the same approach. This is of
particular significance to medical decision in the
medical diagnostic field.
For future work, we intend to add coefficient of
ponderation for each of the five selected features.
We would also to extend our approach to real-world
problems and other public datasets such as heart
disease and breast cancer.
REFERENCES
Kharrat A., Gasmi K., Ben Messaoud M., Benamrane N.,
Abid M., 2010a. Automated Classification of
Magnetic Resonance Brain Images Using Wavelet
Genetic Algorithm and Support Vector Machine. Proc.
9th IEEE Int. Conf. on Cognitive Informatics
(ICCI’10), doi: 10.1109/COGINF.2010.5599712. 369-
-374.
Kharrat A., Ben Messaoud M., Benamrane N., Abid M.,
2010b. Genetic Algorithm for Feature Selection of
MR Brain Images Using Wavelet Co-occurrence.
Proc. IEEE Int. Conf. on Signal and Information
Processing (ICSIP 2010), 1--5.
Boser B. E., Guyon I. M., Vapnik V. N., 1992. A training
algorithm for optimal margin classifiers. Proceedings
of the 5th Annual ACM Workshop on Computational
Learning Theory, 144--152.
Kharrat A., Gasmi K., Ben Messaoud M., Benamrane N.,
Abid M., 2010c. A Hybrid approach for automatic
classification of brain MRI using genetic algorithm
and support vector machine. Leonardo Journal of
Sciences (LJS), 9, 71--82.
Raymer, M. L., Punch, W. F., Goodman, E. D., Kuhn, L.
A., Jain, A. K., 2000. Dimensionality reduction using
genetic algorithms. IEEE Transactions on
Evolutionary Computation, 4, 164--171.
Yang, J., Honavar, V., 1998. Feature subset selection
using a genetic algorithm. IEEE Intelligent Systems,
13, 44--49.
Salcedo-Sanz, S., Prado-Cumplido, M., Pérez-Cruz, F.,
Bousono-Calzon, C., 2002. Feature selection via
genetic optimization. Proceedings of the ICANN
international conference on artificial neural networks,
547--552.
Frohlich, H., Chapelle, O., 2003. Feature selection for
support vector machines by means of genetic
algorithms. Proceedings of the 15th IEEE
international conference on tools with artificial
intelligence, Sacramento, 142--148.
Keith, A. J., Alex, J. B., 1999. The whole brain atlas.
Harvard Medical School, Boston. Available from:
http://med.harvard.edu/AANLIB/.
VISAPP 2011 - International Conference on Computer Vision Theory and Applications
204
