
ARCHITECTURE OF MEDPEER
A New P2P-based System for Integration of Heterogeneous Data Sources
Naïma Souâd Ougouti
1
, Haféda Belbachir
1
, Youssef Amghar
2
and Nabila Aicha Benharkat
2
1
LSSD Laboratory, U.S.T.O LP 1505 El MNaouer, 31000, Oran, Algeria
2
LIRIS UMR 5205, Insa of Lyon, 69620 cedex, Villeurbanne, France
Keywords: Data Mediation, Peer-to-peer Networks, Ontologies, Semantic Web.
Abstract. In this article, we present Medpeer, a new peer-to-peer (P2P) management system for heterogeneous and
distributed data sources. Its principal goal is to provide necessary tools for the semantic mediation of data
from various types (relational, image, text,..) and for the semantic routing of multimodal queries in an P2P
environment. In this environment, each peer will be able to publish the data he wants to share, he is
completely autonomous and the data can belong to different models. MedPeer is a Super-Peer system where
the super-peers are organized by type of data and contain an ontological structure specific to each type.
Each peer exports their data in a common format in the form of a semantically rich ontology in order to
contribute to schemas reconciliation. The queries exchanged have a common format in the form of XML
documents, and are routed towards the relevant peers thanks to a semantic topology built on top of the
existing physical topology.
1 INTRODUCTION
Access to distributed, heterogeneous and
autonomous information sources, has become
possible with the Internet. These information sources
are distinguished by the nature of information,
namely, the ontological domain to which they
belong but also by the type of media they are born
by, such as image, text, video, etc... With the advent
of the semantic web, new opportunities in multi-
sources integration are emerging and many
approaches are revisited, taking into account the new
requirements. We also observe the use or reuse of
datawarehouses, mediators and especially peer-to-
peer systems (Ougouti, 2010).
Recently, several PDMS (Peer Dated
Management Systems) have been born. Senpeer
(Faye, 2006),, Edutella (Nejdl
, 2002), Piazza
(Halevy, 2003), PEPSINT (Cruz, 2004), PeerDB
(Ng, 2003) and Hyperion (Arenas, 2003)
are some
examples of these systems. They combine files
exchange P2P technology such as Napster and
Kaaza with that of distributed databases. They are
based on the semantic description of data sources
that allows also semantic and
intelligent queries
routing and results integration. But, we have noted
that the majority of these systems like Edutella and
PeerDb, treat a maximum of one data model or two
at the same time and do not allow complex and
multimodal queries whose results can be various
types of data like texts, videos and images.
Our objective is to propose solutions to these
problems by presenting a new PDMS: MedPeer. The
principal goal of this system is to provide the
necessary tools for the semantic mediation of
various types of data (relational, image, text,..), the
treatment and semantic routing of multimodal
queries in a P2P environment. In this environment,
each peer will be able to publish the data they want
to share, they are completely autonomous and the
data can belong to different models.
In this article we will only present the
architecture of our system, it is organized as follows:
In section 2 we will present the MedPeer
architecture, and then we will end with a conclusion
and suggest orientations for future work.
2 MEDPEER ARCHITECTURE
2.1 MedPeer Topology
MedPeer has a Super-peer architecture based on
regrouping of peers according to the type of media
351
Ougouti N., Belbachir H., Amghar Y. and Aicha Benharkat N..
ARCHITECTURE OF MEDPEER - A New P2P-based System for Integration of Heterogeneous Data Sources.
DOI: 10.5220/0003661603510354
In Proceedings of the International Conference on Knowledge Management and Information Sharing (KMIS-2011), pages 351-354
ISBN: 978-989-8425-81-2
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)
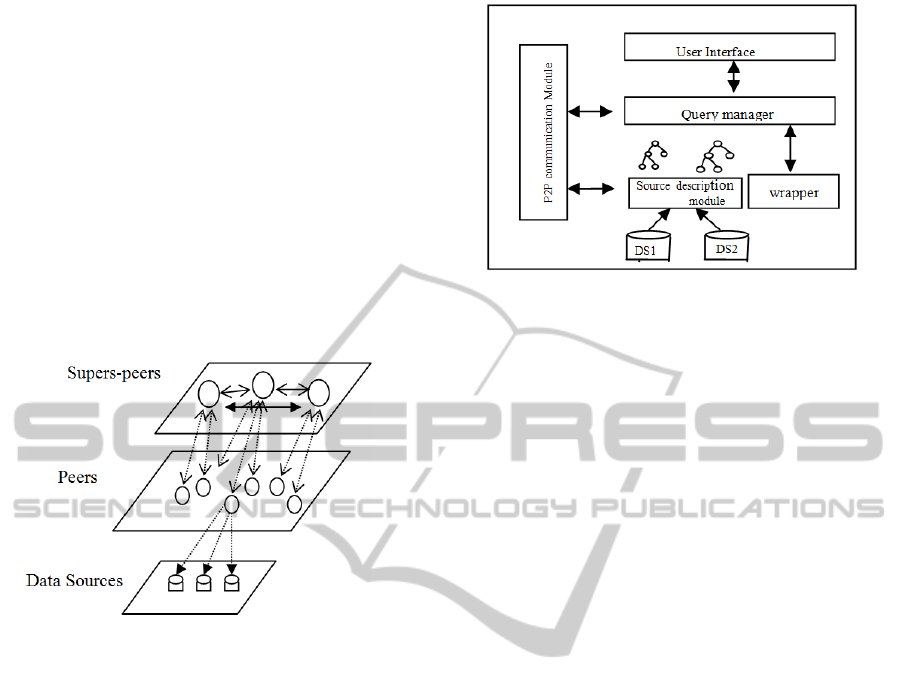
(Texts, Images, Relational databases, semi-
structured,..). This architecture combines a
centralized approach with a non structured one thus
bringing the advantages of centralized research such
as autonomy, and robustness for a distributed
research.
Each super-peer manages the peers containing the
same type of media it is meant to represent; it is
selected according to its calculation capacities and
band-width. In addition, it must have all the
necessary information to be able to direct the
requests arriving to it towards the relevant peers.
The super-peers form between them a pure P2P
network. The peers having different schemas, a
semantic mediation is essential between them.
Figure 1: MedPeer Architecture.
In such a system, to avoid the excessive
translations between peers, there must exist a well
adapted common language; in order to answer this
requirement, we will use an interchange schema
format, based on ontologies, and called structure
ontology. Each super-peer contains a structure
ontology specific to the field it manages. This will
permit semantic schema exchanges between peers
without making assumptions on the data model. A
query interchange format, based on XML allows the
query exchange between peers. In what follows, we
will present in detail the peer and super-peer
components.
2.2 Peer Structure
Each peer has the following components:
Data Source (DS): Each peer is independent
from the others, it contains one or more data
sources which can be relational databases, XML
documents or an images database. The peer
contains its own indexing and research system
by using a suitable, according to the model,
interrogation language (SQL, XQuery, visual,
etc).
Figure 2: Peer Structure.
• Sources Description Module: To regulate the
problem of peers syntactic and semantic
heterogeneity in a community, we use an
ontology as an internal model to represent the
semantic contents of peers. Each data source
present in the peer will be described by an
ontology called lsonto
i
, when i is the source
identifier. These ontologies will be regularly sent
to the super-peer community, to enable it to
generate the semantic correspondences. This also
makes it possible
to deal with the possible
modifications in data sources, then with the
system dynamicity
• Wrapper: This module rewrites the internal
queries into a common exchange format in the
form of an XML document. If the query is
multimodal i.e. returning several types of data in
answer, it will be decomposed by type of data.
Each subquery will be sent to the super-peer
responsible for treating it. This module also
converts the incoming query into the data
model of the local peer.
• Query Manager: Allows the execution of the
local query on the peer and the routing of
subqueries towards the suitable super-peers.
• User Interface: Allows the user to formulate a
local query on its data or a global one on the
network. The queries may refer just to one type
of data and thus carried out within the same
community or to many types of data and thus
carried out through different communities.
• Communication Module: We use JXTA
Open Source platform of Sun to enable the
communication between peers.
2.3 Super-peer Structure
Each super-peer has the following components:
KMIS 2011 - International Conference on Knowledge Management and Information Sharing
352

• Structure Ontology: It is an ontology that
reflects the community data structuring which
the super-peer is responsible. To each type of
data (relational, image..) is associated a structure
ontology that makes it possible to unify the local
concepts used for a semantic reconciliation.
• Mapping Manager: The purpose of this
module is to find all the mappings between data
sources local concepts and those of the structure
ontology thanks to a similarity function which
takes into account the linguistic and semantic
aspects and the various concepts of the semantic
area. The correspondences thus generated will be
stored into an XML document.
• Query Manager: Contains two modules: The
first rewrites the query with the local concepts of
the relevant peers, while the second roots them
towards these same peers. It achieves an
intelligent routing that represents one of the
advantages of the system.
• Network Index: The index contains all
information on the peers of the community and
on all the super-peers of the system. This
information relates to IP address, speed, etc.
• Communication Module: Similar to that of the
peer, based on JXTA platform of Sun.
Figure 3 : Super-peer Structure.
2.4 Ontologies
2.4.1 Structure Ontology
It is an ontology which gathers the whole of the
concepts resulting from the vocabulary used in the
medical field such as the names of relations,
elements or attributes. We propose an ontology
where each concept is defined by its identifier, its
name and its type, it can be connected to other
concepts by certains properties. A property is
defined by its name, its domain and its range, as well
as by its type (aggregation, Composition,
Association, synonym). This ontology is written
with the OWL/RDF language, it takes into account
all types of data defined in the XML Schema
recommendation which provides 44 different types
of data including 19 primitive types and 25 derived
types.
2.4.2 Data Sources Description Ontologies
To facilitate the semantic reconciliation between
peers’ schemas, we describe them thanks to
ontologies. Each handled term in the data sources,
like relation, XML document, attribute or an image
descriptor will be described by the means of a set of
synonyms. In addition, concepts are connected
between them by defined semantic properties
(aggregation, association or composition).
To each concept, a single concept (preferred
term) from the structure ontology will be associated
through the use of a global similarity measurement.
Here is, as an example the diagram of an ontology
describing XML documents.
2.5 Community Creation
When a new super-peer SPj joins the PDMS, it must
present its structure ontology. It announces its
arrival to peers and waits until those among them
that are interested propose their adhesion. This Asp
J
advertisement is in the form of an XML document,
containing the following information: Aspj=(IDSPj,
URIOsj, TDj,
acc
ε
, TTL), in which IDSPj is the
identifier of the super-peer SPj and thus of the
community which it represents, URIOsj represents
the uniform resource identifier of the community
structure ontology, TDj the community data type
(BDR, XML, Texts, Images....),
acc
ε
the minimum
value similarity to accept a mapping between a local
concept and a structure ontology concept. The TTL
(time to live) represents a given delay that stops the
advertisement from buckling
2.6 Peer Adhesion to a Community
When a peer Pi is interested by the super-peer
advertisement, it makes an adhesion request
PiAdh=(IDP, Oli), where IDP is the identifier of the
peer and Oli its local ontology. For each adhesion,
the super-peer index will be will be fed this
information.
The peer will have to give sign of life to the
super-peer before the delay expires. Beyond this
ARCHITECTURE OF MEDPEER - A New P2P-based System for Integration of Heterogeneous Data Sources
353

period if the peer does not manifest itself, it will be
excluded from semantic topology. With its re-
registration, it will have to remake all the known
stages, to take into account possible changes
(addition, suppression, modification) in its structure.
This guarantees a dynamic behavior within the
PDMS, which is strongly desirable in P2P systems.
2.7 Semantic Topology
Nowadays, it has been clearly demonstrated that the
inundation principle in query routing in PDMS
slows down the scale passage. It is thus imperative,
to proceed through a semantic and intelligent
routing.
Semantic topology in MedPeer is built on top of
the physical network, to allow direct queries towards
the relevant peers only. It is built by the super-peer
on the basis of semantic mappings stored within
XML documents.
3 CONCLUSIONS
The current tendency is to revisit the integration
approaches based on mediation and datawarehouses
or to suggest other peer-to-peer systems using the
new possibilities offered by the semantic Web.
The use of ontologies has proved very effective in
semantic integration in the mediators approaches.
But these mediation integration systems are not very
flexible, and the global schema could become a
bottleneck. A strong need, for new decentralized and
dynamic tools is being felt. The peer-to-peer systems
are regarded as a good solution for the Web scale
passage. They present the advantage that they do not
need a single schema, that they allow adding data
and information on the schema of each peer and to
query each peer with its own query language but
they do not handle data semantics. Dealing with
ontologies create a new problem in this field, which
is the definition of semantic mappings between
ontologies in an automatic way.
The MedPeer system that we have presented in
this article takes into account semantics by
describing the sources thanks to ontologies written
with OWL language. The semantic mappings
discovery then becomes easier. The architecture we
propose was conceived with the purpose of dealing
with all types of data such images, videos, texts,
relational data..etc. There is only to define,
beforehand, the specific structure ontology of each
field, or to enrich the one presented in this article.
Our future work will consist in:
• Validating the global similarity function between
two concepts.
• Finding a common queries exchange format
based on XML.
• Defining queries decomposition, rewriting and
routing algorithms.
REFERENCES
Ougouti N. S., Belbachir H., Amghar Y., Benharkat N.,
2010. Integration of Heterogeneous Dated Sources.
Journal of Applied Sciences, 10 (22): (2) 2923-2928,.
Faye D., Nachouki G, Valduriez P., 2006. Integration of
heterogeneous data in SenPeer. ARIMA, Volume
5–1-8
Nejdl W, Wolf B, C qu, Decker S., Sintek Mr., Naeve A.,
Nilsson Mr., Palmér Mr., and Risch T., 2002.
EDUTELLA: With P2P Networking Based
Infrastructure one RDF. In Proceedings of the 11th
International World Wide Web Conference
(WWW2002)
Halevy A. Y., Ives Z G. _ Peter, Mr Tatarinov. I., 2003.
Piazza: Dated Management Infrastructure for
Semantic Web Applications. ACM 1-58113-680-3/
03/0005, Budapest, Hungry
Cruz I F, Xiao H., Hsu F., 2004. Peer-to-Peer Semantic
Integration of XML and RDF Dated Sources. Internal
report, Department of Computer Science, University of
Illinois at Chicago, the USA
Ng W S., Ooi B C, Tan K, and . Zhou A., 2003. PeerDB:
In P2P-based System for Distributed Dated Sharing. In
Proceedings of the 19th International Conference one
Dated Engineering ICDE 633 –644
Arenas Mr., Kantere V, Kementsietsidis A., Kiringa I.,
Miller R. J., and Mylopoulos J., 2003. The Hyperion
Project: From Dated Integration to Data Coordination.
SIGMOD Record32(3):53 –38
KMIS 2011 - International Conference on Knowledge Management and Information Sharing
354
