
USE OF DOMAIN KNOWLEDGE FOR DIMENSION REDUCTION
Application to Mining of Drug Side Effects
Emmanuel Bresso
1,2
, Sidahmed Benabderrahmane
1
, Malika Smail-Tabbone
1
, Gino Marchetti
1
,
Arnaud Sinan Karaboga
1
, Michel Souchet
2
, Amedeo Napoli
1
and Marie-Dominique Devignes
1
1
LORIA UMR 7503, CNRS, Nancy Universit
´
e, INRIA NGE, 54503 Vandoeuvre-les-Nancy, France
2
Harmonic Pharma, Espace Transfert INRIA NGE, 615 Rue du Jardin Botanique, 54600 Villers-les-Nancy, France
Keywords:
Dimension reduction, Clustering, Semantic similarity, Drug side effects.
Abstract:
High dimensionality of datasets can impair the execution of most data mining programs and lead to the pro-
duction of numerous and complex patterns, inappropriate for interpretation by the experts. Thus, dimension
reduction of datasets constitutes an important research orientation in which the role of domain knowledge
is essential. We present here a new approach for reducing dimensions in a dataset by exploiting semantic
relationships between terms of an ontology structured as a rooted directed acyclic graph. Term clustering is
performed thanks to the recently described IntelliGO similarity measure and the term clusters are then used
as descriptors for data representation. The strategy reported here is applied to a set of drugs associated with
their side effects collected from the SIDER database. Terms describing side effects belong to the MedDRA
terminology. The hierarchical clustering of about 1,200 MedDRA terms into an optimal collection of 112 term
clusters leads to a reduced data representation. Two data mining experiments are then conducted to illustrate
the advantage of using this reduced representation.
1 INTRODUCTION AND
MOTIVATION
In some domains such as biology, data complexity
is ubiquitous and constitutes a major challenge for
Knowledge Discovery from Databases (KDD) ap-
proaches. One can anticipate that the more complex
the data, the more relevant and interesting the ex-
tracted knowledge is. However, human expertise is
then heavily required to supervise the KDD process
especially for the data preparation and the interpreta-
tion steps. Thus, an interesting research orientation
consists in exploring how domain knowledge can be
used to help the expert and contribute to the KDD pro-
cess in an automated way.
Complex data are usually voluminous and high-
dimensional. In a classical ObjectsXAttributes repre-
sentation, the number of objects reflects the volume of
data to handle and the number of attributes provides
the number of dimensions in the dataset. Most data
mining methods become rather inefficient when the
dimensionality is too large. Moreover, the patterns ex-
tracted from the data may also reveal inappropriate for
interpretation by the expert due to plethoric, redun-
dant, or non informative attributes. Data reduction is
therefore a crucial issue for data preparation in com-
plex datasets. Several methods exist and are reviewed
in section 2. An interesting situation is the case where
attributes are terms of a controlled and structured vo-
cabulary. In biology and biomedicine such vocabu-
laries (e.g. the Gene Ontology) have been created to
annotate biological objects in databases (genes, pro-
teins, etc.) in order to facilitate the retrieval of ob-
jects sharing similar annotations. Actually, these vo-
cabularies represent basic domain knowledge in the
form of semantic relationships between terms which
enhance subsequent data analyses such as classifica-
tion or characterization.
The present case-study deals with drugs anno-
tated with respect to their side effects. The objects
are drugs retrieved from the Side Effects Repository
(SIDER) database, which compiles all side effects
described in the drug package inserts (Kuhn et al.,
2010). The annotation terms used in SIDER pertain
from the Medical Dictionary for Regulatory Activities
(MedDRA) terminology. Not less than 1,200 Med-
DRA terms are used in SIDER to annotate the drugs.
Dealing with annotation terms taken from a hierar-
chical vocabulary allows to use existing strategies for
attribute reduction such as generalization (Han and
271
Bresso E., Benabderrahmane S., Smail-Tabbone M., Marchetti G., Sinan Karaboga A., Souchet M., Napoli A. and Devignes M..
USE OF DOMAIN KNOWLEDGE FOR DIMENSION REDUCTION - Application to Mining of Drug Side Effects.
DOI: 10.5220/0003662602630268
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2011), pages 263-268
ISBN: 978-989-8425-79-9
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

Kamber, 2001). However, available domain vocabu-
laries ore often organized as a rooted Directed Acyclic
Graph (DAG) rather than a tree structure in which
generalization is intractable. In the present study,
we propose alternatively to cluster annotation terms
based on their similarity in the rooted DAG using
the IntelliGO similarity measure that was initially de-
fined on the Gene Ontology (Benabderrahmane et al.,
2010).
The application of the IntelliGO measure to the
MedDRA vocabulary resulted in the clustering of the
1,200 MedDRA terms into an optimal collection of
112 term clusters. These term clusters led to a reduced
data representation in which drugs are annotated with
term clusters instead of MedDRA terms (Section 3).
Several data mining experiments are then conducted
to show the advantage of using the reduced represen-
tation of the data (Section 4).
2 DATA REDUCTION FOR KDD:
A BRIEF STATE OF THE ART
Methods for data reduction divide into two types: fea-
ture selection and dimension reduction. Feature se-
lection (or attribute selection) is a means of data re-
duction without altering the original data representa-
tion (Guyon and Elisseeff, 2003). Feature selection
methods fall into two categories: filters and wrap-
pers. Filter methods perform feature selection as a
pre-processing step based on a search in the feature
space. Feature subset evaluation relies on measures
that use the training data properties and is carried out
independently of any classifier (John et al., 1994). A
typical example is the correlation-based feature selec-
tion method which eliminates redundant and irrele-
vant attributes by selecting those that individually cor-
relate with the class but have little inter-correlation
(Witten et al., 2011). Concerning the wrapper meth-
ods, they evaluate candidate feature subsets on the ba-
sis of their predictive accuracy (classification perfor-
mance) using a learning algorithm (Kohavi and John,
1997). Most feature selection methods work with su-
pervised classification and use the class information
of the training examples to select the relevant features.
However, Wrapper methods were recently proposed
for feature selection upstream unsupervised learning,
namely clustering (Kim et al., 2000; Dy and Brodley,
2004). Such studies focus on how to evaluate the re-
sults of clustering for each candidate feature subset.
Also,a recent study suggests a filter method indepen-
dent of both the learning algorithm and any predefined
classes by guiding the attribute selection with formal-
ized domain knowledge (Coulet et al., 2008).
Alternative data reduction methods alter the data
representation by encoding the data into a smaller rep-
resentation space. Such dimension reduction is also
called feature compression. The principal compo-
nent analysis is a popular example of such methods
which deals with numeric and continuous data. Clus-
tering was proposed for grouping the attributes in or-
der to improve the classification or the clustering of
textual documents (Kyriakopoulou, 2008). Here the
attributes are binary and correspond to words annotat-
ing textual documents. Word clustering then relies on
comparing their joint distributions in the documents
over the classes (Koller and Sahami, 1996; Slonim
and Tishby, 2000). Thus, the similarity measures used
for clustering the word attributes are corpus-based.
In this paper, we propose that when the attributes
belong to a domain-specific structured vocabulary, a
better clustering of these attributes could be achieved
by using a suitable semantic similarity measure.
3 SEMANTIC CLUSTERING OF
ATTRIBUTES
3.1 The MedDRA Terminology
The MedDRA medical terminology is used to classify
adverse event information associated with the use of
drugs and vaccines. MedDRA is a part of the Uni-
fied Medical Languages System (UMLS) and is of-
ten presented as a hierarchy consisting of five lev-
els: System Organ Class, High Level Group Term,
High Level Term, Preferred Term, Lowest Level Term
(MedDRA, 2007). Lowest level terms correspond
to different terms for the same preferred term. In
the MedDRA terminology, each term has an identi-
fier and all the paths to the root can be download.
For example, for the term C0000733, we have two
paths: C1140263.C0017178.C0947761.C0947846
and C1140263.C0947733.C0021502.C0851837. A
careful review of the parent-child relationships shows
that the MedDRA is actually not a hierarchy: about
37% of the MedDRA terms have more than one di-
rect parent. This together with the natural oriented
of term-term relationship and the absence of cycle,
confers to the MedDRA terminology the status of a
rooted DAG.
3.2 Term-term Semantic Similarity
Measure
Two approaches exist for term-term semantic sim-
ilarity measures: structure-based approaches which
KDIR 2011 - International Conference on Knowledge Discovery and Information Retrieval
272

exploit the structure of the vocabulary (depth, path
length) and corpus-based approaches which exploit
the term distribution in a corpus (annotation fre-
quency, information content). The IntelliGO measure
is a structure-based approach in which the generalized
cosine similarity measure proposed by Ganesan for
hierarchical vocabularies has been adapted to rooted
DAGs (Benabderrahmane et al., 2010). The IntelliGO
measure was initially defined for quantifying simi-
larity between Gene Ontology (GO) terms. For two
terms t
i
, t
j
, it takes into account the maximal depth of
their common ancestors (CA) and the minimal path-
length (SPL) between them:
Sim
IntelliGO
(t
i
,t
j
) =
2MaxDepth(CA)
MinSPL(t
i
,t
j
) + 2MaxDepth(CA)
(1)
To calculate the semantic similarity between two
MedDRA terms, the algorithm starts by retrieving for
each term all their paths to the root node. Then, the
set of common ancestors is defined as the intersec-
tion between the two sets containing the ancestors of
the two terms. In the next step, the algorithm identi-
fies the common ancestors having the maximal depth
from the root node (MaxDepth(CA)). Note that the
Depth of a MedDRA term can be calculated as the
maximal length of a path from this term to the root
node. After that, the algorithm calculates the shortest
path length (MinSPL) separating the two terms. Fi-
nally, the semantic similarity between the two terms
is computed using the equation (1).
As the values of Sim
IntelliGO
range from 0 to 1, we
also define the distance Dist
IntelliGO
as its complement
to 1.
3.3 Clustering MedDRA Terms
A total of 1,288 terms from the 20,037 MedDRA
terms are used in the SIDER database for annotating
drug side effects. Pairwise distances were calculated
for these 1,288 terms. We then used the Ward’s hier-
archical agglomeration algorithm (Ward, 1963) with
an optimization step necessary to select the best level
where to cut the dendrogram in order to obtain a set of
clusters (Kelley et al., 1996). This method defines a
penalty value which is function of the cluster number
and the intra-cluster distance. When this value is min-
imal, the resulting clusters are as highly populated as
possible while simultaneously maintaining the small-
est average intra-cluster distance. In our case the min-
imal penalty value was obtained with 112 clusters
which were subsequently inspected and validated by
two experts. In the rest of this paper, these clusters
will be defined as the term clusters (TC) which will
be used as attributes for data mining.
In order to label each TC with its most represen-
tative term, we introduce a function AvgDist
TC
asso-
ciating to each TC term its average distance to all the
TC terms:
AvgDist
TC
(t
i
) =
1
|TC|
N
TC
∑
j=1
dist(t
i
,t
j
) (2)
Then the representative element R of a TC is the
term which minimizes AvgDist
TC
. For example in
Table 1, Erythema is the representative element of
the TC. The label of a given TC is the concatenation
of the TC number and the representative term (e.g.,
54 Erythema for the TC described in Table 1). Once
TC are built, they can be used for dimension reduc-
tion.
Table 1: Example of TC with the average distance function
calculated for each term t.
Term Cluster Element t AvgDist
T
54
(t)
Decubitus ulcer 0.35
Rash 0.35
Lichen planus 0.32
Parapsoriasis 0.32
Pruritus 0.35
Psoriasis 0.37
Sunburn 0.35
Erythema 0.31
Pityriasis alba 0.32
Photosensitivity reaction 0.37
Rash papular 0.32
Dandruff 0.37
Lupus miliaris disseminatus faciei 0.35
Vulvovaginal pruritus 0.35
4 EVALUATION OF THE IMPACT
OF FEATURE CLUSTERING ON
DATA MINING
4.1 Experimental Design
In order to evaluate the impact of our dimension re-
duction strategy, two data mining experiments were
conducted. The first experiment aims at retrieving fre-
quent associations of side effects shared by drugs in a
given drug category. The second experiment aims at
discriminating drugs belonging to two categories in
terms of side effects. Datasets consist of binary (Ob-
jects X Attributes) relations between drugs (Objects)
and their side effects (Attributes). The side effects are
represented either as individual MedDRA terms or as
TC leading to two data representations.
USE OF DOMAIN KNOWLEDGE FOR DIMENSION REDUCTION - Application to Mining of Drug Side Effects
273

In the first experiment we search for Frequent
Closed Itemsets (FCIs) in order to compare the two
data representations with respect to computation time,
number and relevance of the extracted FCIs. In our
context, a FCI of length n and support s corresponds
to an association of n terms, respectively term clus-
ters, shared by the maximal group of drugs corre-
sponding to a percentage value s of the whole dataset.
The Zart program was used for FCI extraction on the
Coron platform (Szathmary et al., 2007). The experi-
ment was ran on a 2.6GHz processor with 1GB mem-
ory.
As for the second experiment we use the CN2-SD
subgroup discovery algorithm (Lavrac et al., 2004)
with the two data representations in order to check
the impact of term clustering on the computation time
and the produced subgroups. Given a population of
objects and a property of those objects that we are
interested in, subgroup discovery allows to find sub-
groups of objects that are statistically most interest-
ing, i.e., as large as possible and having the most un-
usual distributional characteristics with respect to the
property of interest. In our case, two categories of
drugs are investigated with this method for identify-
ing subgroups of drugs sharing discriminative side-
effects in one category versus the other. The CN2-SD
algorithm implementation used is the one of the Keel
software (Alcala-Fdez et al., 2009) and was executed
on a 8-core 1.86GHz processor with 8GB memory.
4.2 Datasets Description
The category of a drug refers to its therapeutic uses.
The categories of the drugs present in the SIDER
DB are available in the DrugBank DB (Knox et al.,
2011). We chose to perform the data mining experi-
ments on the drugs corresponding to the two largest
categories: the Cardiovascular Agents (CA) and the
Anti-Infective Agents (AIA) containing respectively
94 and 76 drugs.
For each category, we built two datasets : the All
dataset has for attributes the 1,288 MedDRA terms
annotating the drugs in SIDER and the TC dataset has
for attributes the 112 TC (as described in section 3)
note that a TC is assigned to a drug if at least one
member of the TC is reported as annotating the drug
in SIDER. This gives four datasets CA
All
, AIA
All
,
CA
TC
, and AIA
TC
.
4.3 Frequent closed Itemset Extraction
with and without Term Clustering
The Zart program was executed on each dataset with
a minimal support varying from 50 to 100%. Table
Table 2: Number of FCIs for each dataset when varying the
minimal support value.
Minimal
support
50% 60% 70% 80% 90% 100%
AIA
all
178 41 9 2 0 0
AIA
TC
654 154 30 3 0 0
CA
all
386 94 41 11 1 0
CA
TC
5,564 1,379 256 62 6 0
2 summarizes the number of FCIs produced in each
case.
The first observation is the increase in the num-
ber of FCIs for a given minimal support when term
clusters are used. For the AIA
All
and AIA
TC
datasets,
this increase varies from more than 3-fold for mini-
mal supports between 50 and 70% to about 1.5-fold
for higher minimal supports. For CA
All
and CA
TC
datasets this increase goes from about 14-fold at min-
imal support 50 and 60% to 6-fold for higher min-
imal supports. This increase clearly reflects the ex-
pected role of clustering in feature reduction, namely
increasing the density of the binary (Objects X At-
tributes) relation by aggregating object properties.
Accordingly, the computation time of the program
was two-fold longer with the TC than with the All rep-
resentation.
Content analysis of the FCIs was done after rank-
ing FCIs according to their support. The five top-
ranked FCIs (plus ex-aequo) are listed for each dataset
in Figure 1. With the All representation, FCIs can be
very redundant. For example in the AIA
All
dataset
(left panel), three from the five displayed FCIs con-
tain very similar term: nausea, vomiting, nausea and
vomiting symptoms. On the contrary with the TC rep-
resentation (right panel), a unique FCI contains the at-
tribute 64 nausea and vomiting symptoms which rep-
resents the cluster of terms containing all three at-
tributes cited before. Thus, data reduction by term
clustering allows less redundant FCI extraction and
therefore leads to the presentation of more potentially
interesting itemsets to the expert.
Figure 1 also shows that the FCI supports are gen-
erally higher with the TC than with the All data rep-
resentation. A correspondence can be established be-
tween individual terms in the All representation and
the term clusters in the TC representation as illus-
trated in Figure 1 (remember that a term cluster is
labeled with its number and its most representative
term). For example, the {54 Erythema} FCI found
in the AIA
TC
dataset with a support of 88% contains
the term cluster labeled 54
Erythema that includes
the Pruritus individual term and has therefore been
matched with the {Pruritus} FCI found in the AIA
All
dataset with a support of 79%.
KDIR 2011 - International Conference on Knowledge Discovery and Information Retrieval
274
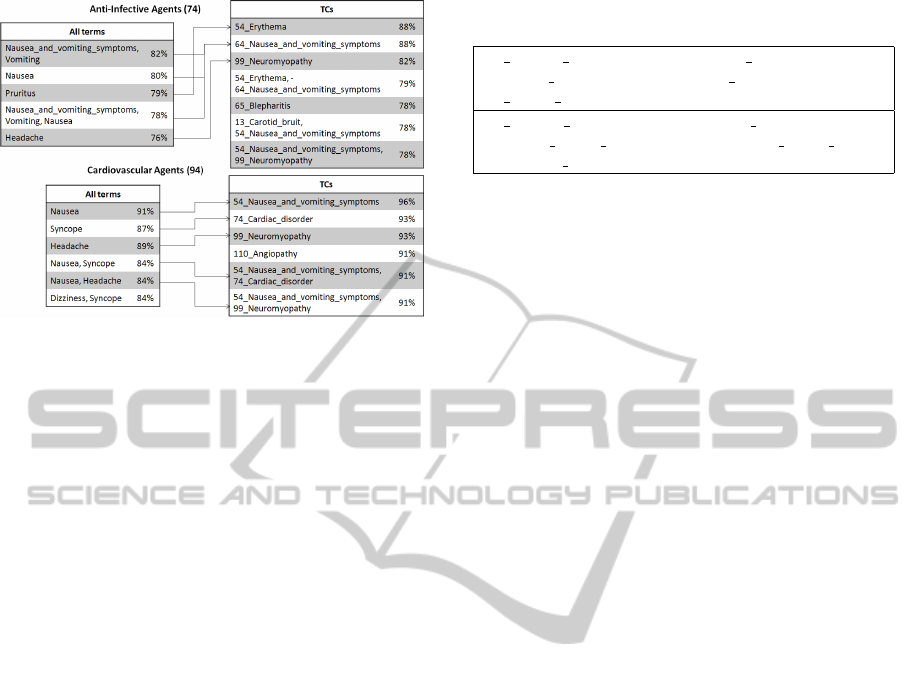
Figure 1: The five most frequent FCIs are extracted from
the total list of FCIs obtained for each dataset. In case of
ex-aequo, all itemsets with the same support are listed.
Furthermore, combined with the lack of redun-
dancy, this increase of FCI support leads to the dis-
covery in the TC datasets of frequent term clusters for
which all individual terms are less frequent and there-
fore not considered in the All datasets.
4.4 Subgroup Discovery with and
without Term Clustering
The second experiment aims at discriminating the
drugs belonging to two categories in terms of pres-
ence or absence of side effects. Indeed, the absence of
side effects may also be important for drug characteri-
zation. This is possible with the CN2-SD algorithm as
the input data is in attribute-value format. We ran the
CN2-SD program on the following unions of datasets
in which an additional attribute was added for the cat-
egory information: CA
All
versus AIA
All
, and CA
TC
versus AIA
TC
.
The first observation concerns the computation
time. When term clusters are used the execution time
is less than ten minutes whereas it does not resume
within six days when all side effects are used. Thus,
data reduction is necessary for successful execution
of the CN2-SD algorithm.
In a second stage, the rules extracted from CA
TC
versus AIA
TC
dataset were analyzed. The left part of a
rule is verified for a number of drugs (support) among
which a certain fraction (coverage) are of the category
indicated in the right part of the rule. The resulting
subgroup therefore identifies a subset of drugs from
this category sharing a specific profile of side effects
with regard to the other category. The best rules in
terms of coverage are shown in Table 3.
To sum up, these results show that the data reduc-
Table 3: Best rules (with coverage / support) extracted by
the CN2-SD program for CA
TC
versus AIA
TC
.
50 Angina pectoris = T AND 93 Bacteraemia = F
AND 52 Ichthyosis = F AND 54 Erythema = T AND
49 Folate deficiency = F => CA (0.96/56)
31 Splenic infarction = T AND 41 Neutropenia = T
AND 42 Penile discharge = F AND 77 Facial pain =
T AND 79 Cachexia = T => AIA (0.88/26)
tion used allows subgroup discovery which was im-
possible with the extended data representation. Fur-
ther investigation by domain experts is ongoing.
5 DISCUSSION AND
PERSPECTIVES
In this paper we have reported a method for dimension
reduction guided by domain knowledge. The method
is based on attribute clustering using a semantic sim-
ilarity measure. We took advantage of our recently
defined IntelliGO similarity measure which applies to
the rooted DAG structure encountered in many vocab-
ularies.We believe that our strategy can be applied in
various other biomedical context (Leva et al., 2005;
Pakhomov et al., 2007). We tested our method on
a dataset of 170 drugs annotated with 1,288 terms
taken from the MedDRA terminology and represent-
ing the drugs possible side effects. Using IntelliGO-
based term-term distances and hierarchical clustering,
we reduced data representation from 1,288 individ-
ual terms down to 112 term clusters. In this work we
adopted a binary representation for the reduced data
representation, i.e., a TC is assigned to a drug if at
least one of its elements has been associated with the
drug in the SIDER database. This representation ig-
nores the impact of multiple associations between a
drug and TC elements. A many-valued relation could
be produced to take into account such situations. Re-
cently described extension of formal concept analysis
may help us handling such data representation (Mes-
sai et al., 2008; Kaytoue-Uberall et al., 2009).
The dimension reduction method we have devel-
oped was tested with two data mining algorithms: FCI
extraction and subgroup discovery. The results show
that FCIs extracted from the TC data representation
are less redundant and display higher supports than
from the All representation. Another consequence of
the data reduction is that the expert’s task is facili-
tated because more relevant and explicit side effects
are found among FCIs displaying high support. As
for subgroup discovery, dimension reduction revealed
to play a crucial role. Indeed, the program was unable
to resume with the All data representation whereas it
USE OF DOMAIN KNOWLEDGE FOR DIMENSION REDUCTION - Application to Mining of Drug Side Effects
275

provided, with the reduced TC representation, quite
interesting rules characterizing subgroups of one drug
category versus another one. Complementary experi-
ments can now be carried out to identify rules specific
of a given category versus all other categories.
REFERENCES
Alcala-Fdez, J., Snchez, L., Garca, S., del Jesus, M., Ven-
tura, S., Garrell, J., Otero, J., Romero, C., Bacardit,
J., Rivas, V., Fernndez, J., and Herrera, F. (2009).
KEEL: a software tool to assess evolutionary algo-
rithms for data mining problems. Soft Computing -
A Fusion of Foundations, Methodologies and Appli-
cations, 13(3):307–318–318.
Benabderrahmane, S., Smail-Tabbone, M., Poch, O.,
Napoli, A., and Devignes, M. (2010). IntelliGO: a new
vector-based semantic similarity measure including
annotation origin. BMC Bioinformatics, 11(1):588.
Coulet, A., Smail-Tabbone, M., Benlian, P., Napoli, A., and
Devignes, M. (2008). Ontology-guided data prepa-
ration for discovering genotype-phenotype relation-
ships. BMC Bioinformatics, 9(Suppl 4):S3.
Dy, J. G. and Brodley, C. E. (2004). Feature selection for
unsupervised learning. J. Mach. Learn. Res., 5:845–
889.
Guyon, I. and Elisseeff, A. (2003). An introduction to
variable and feature selection. J. Mach. Learn. Res.,
3:1157–1182.
Han, J. and Kamber, M. (2001). Data Mining: Concepts
and Techniques. Morgan Kaufmann, San Francisco, 1
edition.
John, G. H., Kohavi, R., and Pfleger, K. (1994). Irrelevant
Features and the Subset Selection Problem. In In-
ternational Conference on Machine Learning, pages
121–129.
Kaytoue-Uberall, M., Duplessis, S., Kuznetsov, S. O., and
Napoli, A. (2009). Two fca-based methods for mining
gene expression data. In ICFCA, pages 251–266.
Kelley, L. A., Gardner, S. P., and Sutcliffe, M. J. (1996).
An automated approach for clustering an ensemble
of NMR-derived protein structures into conforma-
tionally related subfamilies. Protein Engineering,
9(11):1063 –1065.
Kim, Y., Street, W. N., and Menczer, F. (2000). Feature
selection in unsupervised learning via evolutionary
search. In Proceedings of the sixth ACM SIGKDD in-
ternational conference on Knowledge discovery and
data mining, pages 365–369, Boston, Massachusetts,
United States. ACM.
Knox, C., Law, V., Jewison, T., Liu, P., Ly, S., Frolkis, A.,
Pon, A., Banco, K., Mak, C., Neveu, V., Djoumbou,
Y., Eisner, R., Guo, A. C., and Wishart, D. S. (2011).
DrugBank 3.0: a comprehensive resource for Omics
research on drugs. Nucleic Acids Research, 39(suppl
1):D1035 –D1041.
Kohavi, R. and John, G. H. (1997). Wrappers for feature
subset selection. Artificial Intelligence, 97(1-2):273–
324.
Koller, D. and Sahami, M. (1996). Toward optimal feature
selection. In Saitta, L., editor, Proceedings of the Thir-
teenth International Conference on Machine Learning
(ICML), pages 284–292. Morgan Kaufmann Publish-
ers.
Kuhn, M., Campillos, M., Letunic, I., Jensen, L. J., and
Bork, P. (2010). A side effect resource to capture phe-
notypic effects of drugs. Mol Syst Biol, 6.
Kyriakopoulou, A. (2008). Text classification aided by clus-
tering: a literature review. In Tools in Artificial Intel-
ligence, chapter 14. Paula Fritzsche, intech edition.
Lavrac, N., Kavsek, B., Flach, P., and Todorovski, L.
(2004). Subgroup discovery with CN2-SD. J. Mach.
Learn. Res., 5:153–188.
Leva, A. D., Berchi, R., Pescarmona, G., and Sonnessa,
M. (2005). Analysis and prototyping of biological
systems: the abstract biological process model. In-
ternational Journal of Information and Technology,
3(4):216–224.
MedDRA (2007). Meddra maintenance and support ser-
vices organization. introductory guide, meddra ver-
sion 10.1.
Messai, N., Devignes, M.-D., Napoli, A., and Sma
¨
ıl-
Tabbone, M. (2008). Many-valued concept lattices
for conceptual clustering and information retrieval. In
ECAI, pages 127–131.
Pakhomov, S. S., Hemingway, H., Weston, S. A., Jacob-
sen, S. J., Rodeheffer, R., and Roger, V. L. (2007).
Epidemiology of angina pectoris: Role of natural lan-
guage processing of the medical record. American
Heart Journal, 153(4):666–673.
Slonim, N. and Tishby, N. (2000). Document clustering
using word clusters via the information bottleneck
method. In Proceedings of the 23rd annual interna-
tional ACM SIGIR conference on Research and de-
velopment in information retrieval, pages 208–215,
Athens, Greece. ACM.
Szathmary, L., Napoli, A., and Kuznetsov, S. O. (2007).
Zart: A multifunctional itemset mining algorithm. In
CLA, pages 26–37.
Ward, J. H. (1963). Hierarchical grouping to optimize
an objective function. Journal of the American Sta-
tistical Association, 58(301):236–244. ArticleType:
research-article / Full publication date: Mar., 1963 /
Copyright 1963 American Statistical Association.
Witten, I. H., Frank, E., and Hall, M. A. (2011). Data
Mining: Practical Machine Learning Tools and Tech-
niques. Morgan Kaufmann, Burlington, MA, 3 edi-
tion.
KDIR 2011 - International Conference on Knowledge Discovery and Information Retrieval
276
