
SEMI-SUPERVISED EVALUATION OF CONSTRAINT SCORES
FOR FEATURE SELECTION
Mariam Kalakech
1,3
, Philippe Biela
1
, Denis Hamad
2
and Ludovic Macaire
3
1
HEI, 13 Rue de Toul, F-59046, Lille, France
2
LISIC, ULCO, 50 Rue Ferdinand Buisson, F-62228, Calais, France
3
LAGIS FRE CNRS 3303, Universit´e Lille 1, Bˆatiment P2, Cit´e Scientifique, F-59655, Villeneuve d’Ascq, France
Keywords:
Feature selection, Constraint scores, Pairwise constraints, Semi-supervised evaluation.
Abstract:
Recent feature constraint scores, that analyse must-link and cannot-link constraints between learning samples,
reach good performances for semi-supervised feature selection. The performance evaluation is generally based
on classification accuracy and is performed in a supervised learning context. In this paper, we propose a
semi-supervised performance evaluation procedure, so that both feature selection and classification take into
account the constraints given by the user. Extensive experiments on benchmark datasets are carried out in the
last section. They demonstrate the effectiveness of feature selection based on constraint analysis.
1 INTRODUCTION
In machine learning and pattern recognition appli-
cations, the processing of high dimensional data re-
quires large computation time and capacity storage.
Though, it leads to poor performances when the di-
mensionality to sample size ratio is high. To im-
prove performances of data classification, the sample
dimensionality is reduced thanks to a feature selec-
tion scheme. It consists in selecting the most relevant
features in order to build a low dimensional feature
space. One generally assumes that a classifier scheme
operating in this low dimensional feature space out-
performs the same classifier operating in the initial
feature space
The feature subspace can be selected thanks to a
non-exhaustive sequential feature selection procedure
which iteratively adds selected features (Kudo and
Sklansky, 2000). However, such a strategy is time
consuming since it evaluates properties of different
multi-dimensional sub-spaces. That leads authors to
sort the score of each feature, so that the feature sub-
space is composed of the most relevant ones (Liu and
Motoda, 1998).
During the training step, the score of each feature
is evaluated thanks to the subset of training samples.
They can be either unlabelled or labelled, leading to
the development of unsupervised and supervised fea-
ture selection techniques. Unsupervised feature score
measures the feature ability of keeping the intrinsic
data structure. In the supervised learning context, the
feature score is based on the correlation between the
feature and the class labels of the training samples.
However, in the supervised learning context, the
sample labelling process achieved by the user is fas-
tidious and expensive. That is the reason why for
many real applications, the training data subset is
composed of a few labelled samples and huge unla-
belled ones. To deal with this ’lack labelled-sample
problem’, recent semi-supervised feature scores have
been developed (Zhao and Liu, 2007),(Zhao et al.,
2008).
Beside class labels of samples, there is another
kind of user supervision information called the pair-
wise constraints. The user simply specifies whether
a pair of training samples must be regrouped to-
gether (must-link constraints) or cannot be regrouped
together (cannot-link constraints). Recent feature
scores called constraint scores, that analyse must-link
and cannot link constraints, have shown excellent per-
formance of semi-supervised learning with a lot of
datasets (Zhao et al., 2008),(Zhang et al., 2008).
To measure the performances reached by feature
selection schemes based on constraint scores, authors
use benchmarkdatasets composed of labeled samples.
Each dataset is divided into the training and the test
subsets according to the holdout strategy. A small
number of must-link and cannot-link constraints are
deduced from the labeled samples of the training sub-
set. Finally, the training subset is only composed of
175
Kalakech M., Biela P., Hamad D. and Macaire L..
SEMI-SUPERVISED EVALUATION OF CONSTRAINT SCORES FOR FEATURE SELECTION.
DOI: 10.5220/0003680001750182
In Proceedings of the International Conference on Neural Computation Theory and Applications (NCTA-2011), pages 175-182
ISBN: 978-989-8425-84-3
Copyright
c
2011 SCITEPRESS (Science and Technology Publications, Lda.)

constrained and unconstrained samples without any
knowledge about their label. First, the features are se-
lected by sorting their constraint scores obtained with
the training samples. Then, the performance of the
feature selection algorithm based on each constraint
score is measured by the classification accuracy of test
samples reached by a classifier operating in the fea-
ture space defined by the selected features. The near-
est neighbor classifier is the most used for this pur-
pose. As it requires a lot of prototypes of the classes,
it uses training samples with their labels as proto-
types whereas these labels have not been exploited
by the constraint scores. Indeed, a constraint score
only analyses unconstrained data samples and/or a
few pairwise constraints. So, thetest samples are clas-
sified in a supervised learning context (the prototypes
are the training samples with their labels) whereas
the features are selected in a semi-supervised learn-
ing context (only constraints on a few training sam-
ples are considered).
In this paper, we propose a semi-supervised evalu-
ation procedure of performance reached with the con-
straint scores, so that both feature selection and test
sample classification take into account the constraints
given by the user.
The paper is organized as follows. Constraint
scores used by semi-supervised feature selection
schemes, are introduced in section 2. In section 3, we
describe our semi-supervised procedure of constraint
score evaluation. Finally, experiments presented in
section 4, compare the performances of the different
constraint scores thanks to the semi-supervised eval-
uation.
2 CONSTRAINT SCORES
Given the training subset composed of n samples de-
fined in a d-dimensional feature space, let us denote
X = (x
ir
) i = 1, .. .,n; r = 1, .. .,d; the associated
data matrix where x
ir
is the r
th
feature value of the
i
th
data. Each of the n rows of the matrix X rep-
resents a data sample x
i
= (x
i1
,. .. ,x
id
) ∈ R
d
, while
each of the d columns of X defines the feature values
f
r
= (x
1r
,. .. ,x
nr
)
T
∈ R
n
.
2.1 Constraints
In the semi-supervised learning context, the prior
knowledge about the data is usually represented by
sample labels. Furthermore, another kind of knowl-
edge is represented by pairwise constraints. Pairwise
constraints simply mention for some pairs of data
samples that they are similar, i.e. must be grouped to-
gether (must-link constraints), or that they are dissim-
ilar, i.e. cannot be grouped together (cannot-link con-
straints). These pairwise constraints arise naturally in
many applications since they are easier to be obtained
by the user than the class labels. They simply formal-
ize that two data samples belong or not to the same
class without detailed information about the different
classes.
The user has to build the subset M of must-link con-
straints and the subset C of cannot-link constraints de-
fined as:
M =
(x
i
,x
j
), such as x
i
and x
j
must be linked
,
C =
(x
i
,x
j
), such as x
i
and x
j
cannot be linked
.
The cardinals of these subsets are usually much
lower than the number n(n− 1)/2 of all possible pair-
wise constraints defined by the data.
In the context of the spectral theory, two specific
graphs are built from these two subsets:
• The must-link graph G
M
where a connection is
established between two nodes i and j if there is a
must-link constraint between their corresponding
samples (nodes) x
i
and x
j
.
• The cannot-link graph G
C
where two nodes i and
j are connected if there is a cannot-link constraint
between their corresponding samples (nodes) x
i
and x
j
.
The connection weights between two nodes of the
graphs G
M
and G
C
are respectivelystored by the sim-
ilarity matrices S
M
(n × n) and S
C
(n × n), and are
built as:
s
M
ij
=
1 if (x
i
,x
j
) ∈ M or (x
j
,x
i
) ∈ M
0 otherwise,
(1)
s
C
ij
=
1 if (x
i
,x
j
) ∈ C or (x
j
,x
i
) ∈ C
0 otherwise.
(2)
2.2 Scores
This prior knowledge represented by the constraints
has been integrated in many recent feature scores
(Zhang et al., 2008) (Zhao et al., 2008) (Kalakech
et al., 2011).
Zhang et al. propose two constraint scores C
1
r
and
C
2
r
which use only the subset of must-link and cannot-
link constraints (Zhang et al., 2008):
C
1
r
=
∑
i
∑
j
(x
ir
− x
jr
)
2
s
M
ij
∑
i
∑
j
(x
ir
− x
jr
)
2
s
C
ij
=
f
T
r
L
M
f
r
f
T
r
L
C
fr
, (3)
C
2
r
=
∑
i
∑
j
(x
ir
− x
jr
)
2
s
M
ij
− λ
∑
i
∑
j
(x
ir
− x
jr
)
2
s
C
ij
= f
T
r
L
M
f
r
− λ f
T
r
L
C
f
r
,
(4)
NCTA 2011 - International Conference on Neural Computation Theory and Applications
176

where L
M
= D
M
− S
M
and L
C
= D
C
− S
C
, are the
constraint Laplacian matrices, D
M
and D
C
are the de-
gree matrices defined by D
M
ii
= d
M
i
(d
M
i
=
∑
n
j=1
s
M
ij
)
and D
C
ii
= d
C
i
( d
C
i
=
∑
n
j=1
s
C
ij
) and λ is a regulariza-
tion coefficient used to balance the contribution of
must-link and cannot-link constraints. Must-link con-
straints are favored by setting 0 < λ < 1 and the lower
these two scores are, the more efficient the feature is.
Zhao et al. define another score which uses both
unconstrained data and pairwise constraints in order
to retrieve both locality properties and discriminat-
ing structures of the data samples (Zhao et al., 2008).
They build a new graph G
W
that connects samples
having high probability of sharing the same label:
• G
W
is the within-class graph: two nodes i and j
are connected if (x
i
,x
j
) or (x
j
,x
i
) belongs to M ,
or if one of the two samples is unconstrained but
they are sufficiently close to each other (by using
the k-nearest neighbor graph denoted kNN)
The edges in the graph G
W
are weighted by using the
similarity matrix S
W
(n× n) expressed as:
s
W
ij
=
γ if (x
i
,x
j
) ∈ M or (x
j
,x
i
) ∈ M
1 if x
i
or x
j
is unlabeled
but x
i
∈ kNN(x
j
) or x
j
∈ kNN(x
i
)
0 otherwise
(5)
where γ is a suitable constant parameter which has
been empirically set to 100 in (Zhao et al., 2008).
Zhao et al. also introduce a Laplacian score, called
the locality sensitive discriminant analysis score and
defined as:
C
3
r
=
∑
i
∑
j
(x
ir
− x
jr
)
2
s
W
ij
∑
i
∑
j
(x
ir
− x
jr
)
2
s
C
ij
=
f
T
r
L
W
f
r
f
T
r
L
C
f
r
. (6)
where L
W
= D
W
− S
W
, D
W
being the degree matrix
defined by D
W
ii
= d
W
i
(d
W
i
=
∑
n
j=1
s
W
ij
). The lower
the score C
3
is, the more relevant the feature is.
The scores C
1
and C
2
do not take into account the
unconstrained samples since they are only based on
the must-link and cannot link constraints. C
3
consid-
ers mainly the must-link constraints, so it seems to be
very close to C
2
and both neglect the unconstrained
samples.
Though, taking into account the unconstrained
samples should catch the data structure and make
less sensitive a feature score against the given con-
straint subsets. That is why we have proposed a semi-
supervised constraint score C
4
defined as (Kalakech
et al., 2011):
C
4
r
=
˜
f
T
r
L
˜
f
r
˜
f
T
r
D
˜
f
r
.
f
T
r
L
M
f
r
f
T
r
L
C
f
r
, (7)
where D and L are respectively the degree and the
Laplacian matrices (L = D−S) deduced from the sim-
ilarity matrix S. S (n× n) is the similarity matrix be-
tween all the samples, expressed as:
s
ij
= exp
−
x
i
− x
j
2
2t
2
!
. (8)
t is a Gaussian parameter ajusted by the user.
The score C
4
r
is the simple product between the
unsupervised Laplacian score processed with samples
(He et al., 2005) and the constraint score C
1
r
(see
Equation (3)) (Zhang et al., 2008). As for the other
scores, the features are ranked in ascending order ac-
cording to score C
4
in order to select the most relevant
ones. We have experimentally demonstrated that this
score is less sensitive to the constraint changes than
the classical scores while selecting features with com-
parable classification performances (Kalakech et al.,
2011) (Kalakech et al., 2010).
3 EVALUATION SCHEME
In this section, we present the classical supervised
evaluation scheme used to evaluate the performances
of the features selected by the constraint scores, and
propose our new semi-supervised evaluation scheme.
3.1 Supervised Evaluation
In order to compare different feature scores, the
dataset is divided into training and test subsets. The
feature selection procedure is performed with the
training subset. Then, the performance of each con-
straint score is measured by the accuracy rates of test
sample obtained by a classifier such as kNN classifier
operating in the feature space defined by the selected
features.
The training samples with their true labels are
used by the nearest neighbor classifier to classify the
test data, whereas these true labels have not been
used by the constraint scores. Indeed, these scores
use only the unconstrained data and/or a few pairwise
constraints given by the user. So, the test samples
are classified in the supervised learning context (the
prototypes are the training samples with their true la-
bels) whereas the features are selected in the semi-
supervised learning context (only constraints on a few
training samples are analyzed).
Though, the selection and the evaluation should
operate in the same learning context. That is why,
unlike to classical supervised evaluation, we propose
to perform the score evaluation in a semi-supervised
context.
SEMI-SUPERVISED EVALUATION OF CONSTRAINT SCORES FOR FEATURE SELECTION
177

Algorithm 1: Constrained K-means.
1: Choose randomly K samples as the initial class
centers.
2: Assign each sample to the closet class while ver-
ifying that constraint subsets M and C are not
violated.
3: Update the center of each class.
4: Iterate 2 and 3 until converge.
3.2 Semi-supervised Evaluation
To compare the performances reached by differ-
ent feature selection schemes operating in the semi-
supervised learning context, the test samples are also
classified in the semi-supervised context. For this pur-
pose, feature selection and test sample classification
take into account only the constraints given by the
user as prior knowledge. To classify the test samples,
the nearest neighbor classifier needs to define the la-
bels of the training samples.
In the semi-supervised context, we have no prior
knowledge about labels of these training samples. So,
to build the prototypes of classes, we propose to esti-
mate the labels of the training samples. As the prior
knowledge is described by a few constraints between
training samples, we propose to cluster the training
samples thanks to the constrained K-means scheme
developed by Wagstaff et al. (Wagstaff et al., 2001)
(see Algorithm 1). The desired number of classes is
set by the user and this scheme operates in the selected
features space.
Once the labels of the training samples have been
estimated, the nearest neighbor classifier use them as
prototypes of classes to classify the test samples.
Since the true classes of the training samples can
be different from those determined by the constrained
K-means, we cannot directly use these labels to mea-
sure the classification accuracy of the test samples.
For this purpose, we propose to match the true and
estimated labels of the training samples thanks to
Carpaneto and Toth algorithm (Carpaneto and Toth,
1980).
4 EXPERIMENTS
In this section, we compare the different constraint
scores performances thanks to the semi-supervised
evaluation. Experiments are achieved with six well
known and largely used benchmark databases, and
more precisely the ’Wine’, ’Image segmentation’
and ’Vehicle’ databases from the UCI repository
((Blake et al., 1998)), the face database ’ORL’
((Samaria and Hartert, 1994)) and the two gene ex-
pression databases, i.e., ’Colon Cancer’((Alon et al.,
1999)) and ’Leukemia’((Golub et al., 1999)). These
databases have been retained since the features are nu-
meric and since the label information of each sample
is clearly defined.
4.1 Datasets
In our experiments, we first normalize the features be-
tween 0 and 1, so that the scale of the different fea-
tures is the same. For each dataset, we follow a Hold-
out partition and choose the half of samples from each
class as the training data and the remaining data for
testing.
Here is a brief description of the six considered
databases:
• ’Wine’ Database
This database contains 178 samples characterized
by 13 features (d=13) composed of 3 classes hav-
ing 59, 71 and 48 instances, respectively. We ran-
domly select 30, 36 and 24 samples from each
class to build the training subset. The remaining
samples are considered as the test subset.
• ’Image Segmentation’ Database
This database contains 210 samples characterized
by 19 features (d=19) regrouped into 7 classes,
each class having 30 instances. We randomly se-
lect 15 samples from each class to build the train-
ing subset and the remaining samples constitute
the test subset.
• ’Vehicle’ Database
This database contains 846 samples characterized
by 18 features (d=18) regrouped into 4 classes,
having 212, 217, 218 and 199 instances, respec-
tively. We randomly select 106, 109, 109 and 100
samples from each class to build the training sub-
set.
Figure 1: Sample face images from the ORL database (2
subjects).
The ’ORL’ database (Olivetti Research Labora-
tory) contains a set of face images representing
40 distinct subjects. There are 10 different im-
ages per subject, so that the database contains 400
NCTA 2011 - International Conference on Neural Computation Theory and Applications
178

images. For each subject, the images have been
acquired according to different conditions: light-
ing, facial expressions (open / closed eyes, smil-
ing / not smiling) and facial details (glasses / no
glasses) (see Figure 1).
In our experiments, original images are normal-
ized (in scale and orientation) so that the two eyes
are aligned at the same horizontal position. Then,
the facial areas are cropped in order to build im-
ages of size 32 × 32 pixels, whose gray level is
quantified with 256 levels. Thus, each image
can be represented by a 1024-dimensional sam-
ple data.
We randomly select 5 images from each class
(subject) to build the training subset. The remain-
ing samples are organized as the test subset.
• ’Colon Cancer’ database
This database contains 62 tissues (40 tumors and
22 normals) characterized by the expression of
2000 genes. We randomly select 20 and 11 sam-
ples from each class to build the training subset.
The remaining data are organized as the test sub-
set.
• ’Leukemia’ database
This database contains information on gene-
expression in samples from human acute myeloid
(AML) and acute lymphoblastic leukemias
(ALL). From the originally measured 6817 genes,
the genes that are not measured in at least one
sample, are removed. So a total of 5147 genes are
examined in the experiments.
Because Leukemia has a predefined partition of
the data into training (27 ALL and 11 AML) and
test (20 ALL and 14 AML) subsets ((Golub et al.,
1999)), all the experiments on this dataset are
performed on these predefined training and test
subsets.
4.2 Experimental Procedure
In our experiments, the feature selection is performed
on the training samples and features are ranked ac-
cording to the different scores. At each feature se-
lection run q, q=1,..., p, we simulate the generation
of pairwise constraints as follow: we randomly se-
lect pairs of samples from the training subset and cre-
ate must-link or cannot-link constraints depending on
whether the underlying classes of the two samples are
the same or different. We iterate this scheme until we
obtain l must-link constraints and l cannot-link con-
straints, l being set by the user.
The classification accuracies of the test samples
are used to evaluate the performance of each score.
The rates of good classification are averaged over p =
100 runs with different generations of constraints. 10
constraints (l=5) have been considered for the ’Wine’,
’Image segmentation’, ’Vehicle’ and ’ORL’ databases
and 60 constraints (l=30) have been consideredfor the
’Colon Cancer’ and ’Leukemia’ ones.
4.3 Accuracy vs. Number of Features
We compare the accuracy rates of the different scores
C
1
, C
2
, C
3
and C
4
thanks to our semi-supervised eval-
uation procedure. The labels of the training data are
estimated by the constrained K-means algorithm op-
erating in the selected feature space. These estimated
labels are then used by the nearest neighbor classi-
fier in order to measure the accuracy of the different
scores.
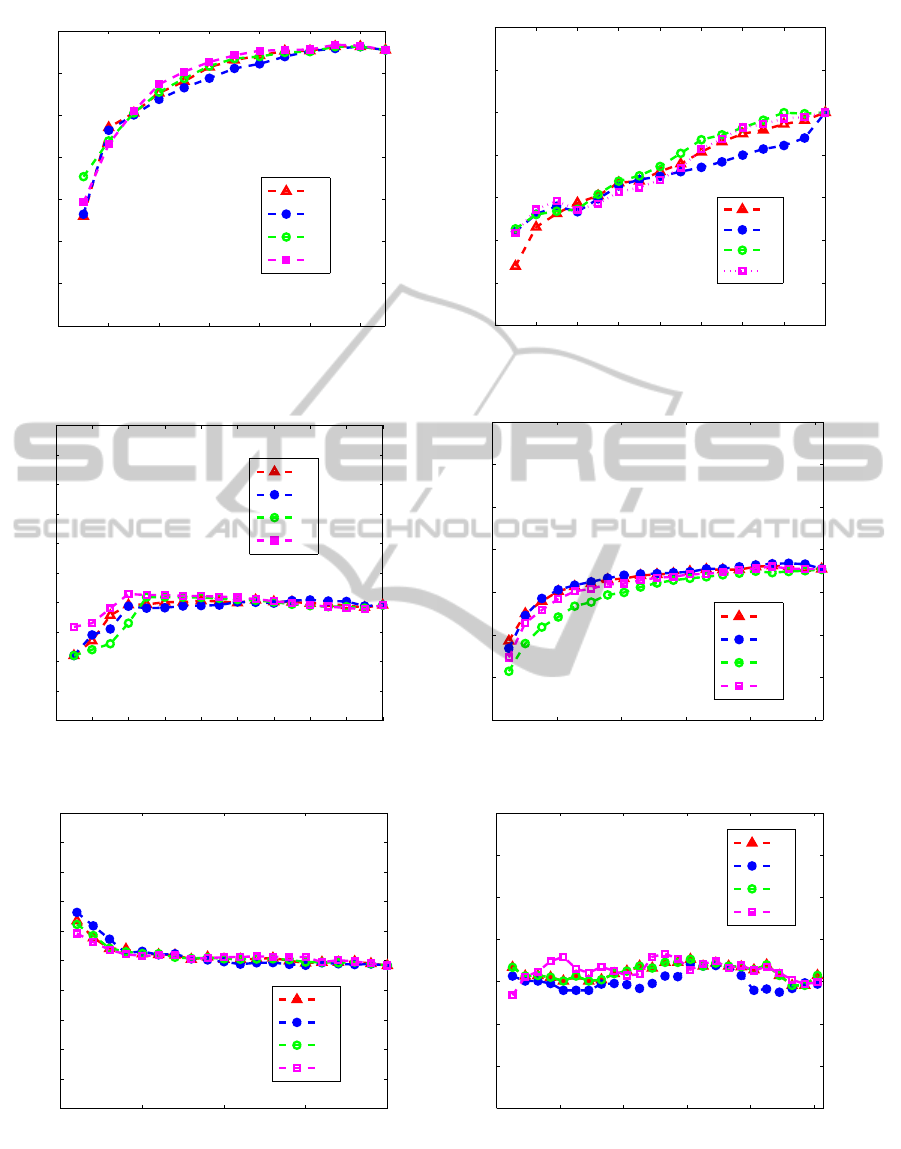
Figure 2 shows the accuracy rates vs the de-
sired number of selected features on the databases
of ’Wine’,’Image segmentation’, ’Vehicle’, ’ORL’,
’Colon cancer’ and ’Leukemia.
From this figure, we can see that the accuracy rates
of C
1
, C
2
, C
3
and C
4
are very close, because the dif-
ferent curves overlap. Since these results are averaged
over 100 runs, it is hard to compare them.
That leads us to compare these scores by exam-
ining their accuracies at each of the 100 runs. For a
fixed number of selected features, in each of the 100
runs, we propose to rank the 4 scores in descending
order of their accuracy.
Let us denote rank
∗
q
the rank of the criterion C
∗
at the run q. This rank takes the values 1, 2, 3 or 4.
At each run q, the score having the highest accuracy
is ranked as 1 and the score with the lowest accuracy
value, is ranked as 4. Scores with the same accuracy
have the same rank.
We calculate the rank sum T
∗
for each semi-
supervised constraint score as follow:
T
∗
=
100
∑
q=1
rank
∗
q
, (9)
where * is 1, 2, 3 or 4 corresponding to the score C
1
,
C
2
, C
3
or C
4
respectively. The method with the low-
est rank sum is considered as being the score which
provides the best results.
Table 1 shows the rank sum T
∗
for the different
databases ’Wine’, ’Image segmentation’, ’Vehicle’,
’ORL’, ’Colon Cancer’ et ’Leukemia’. The rank sum
of each score is computed by considering respectively
the first 6, 5, 8, 300, 1000 and 2576 features on the
’Wine’, ’Image segmentation’, ’Vehicle’ and ’ORL’
databases.
Our score C
4
has the lowest value 3 times (indi-
cated as bold) over the 6 rows of table 1. The other
three scores share each one a row.
SEMI-SUPERVISED EVALUATION OF CONSTRAINT SCORES FOR FEATURE SELECTION
179
0 2 4 6 8 10 12
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Number of selected features
Accuracy
C
1
C
2
C
3
C
4
(a) ’Wine’.
0 2 4 6 8 10 12 14 16
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Number of selected features
Accuracy
C
1
C
2
C
3
C
4
(b) ’Image segmentation’.
0 2 4 6 8 10 12 14 16 18
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Number of selected features
Accuracy
C
1
C
2
C
3
C
4
(c) ’Vehicle’.
0 200 400 600 800 1000
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Number of selected features
Accuracy
C
1
C
2
C
3
C
4
(d) ’ORL’.
0 500 1000 1500 2000
0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Number of selected features
Accuracy
C
1
C
2
C
3
C
4
(e) ’Colon Cancer’.
0 1000 2000 3000 4000 5000
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
Number of selected features
Accuracy
C
1
C
2
C
3
C
4
(f) ’Leukemia’.
Figure 2: Accuracy rates vs. the desired number of selected features on the 6 databases. 10 constraints composed of 5
must-link et 5 cannot-link was used for the ’Wine’, ’Image segmentation’, ’Vehicle’ and ’ORL’ databases and 60 constraints
composed of 30 must-link and 30 cannot-link was used for the ’Colon Cancer’ and ’Leukemia’ databases. The evaluation is
performed in a semi-supervised learning context: the one nearest neighbor classifier uses the estimated labels of the training
samples as prototypes of classes.
NCTA 2011 - International Conference on Neural Computation Theory and Applications
180

Table 1: The rank sum of the constraint scores on different
databases.
Database \T T
1
T
2
T
3
T
4
’Wine’ 191 277 206 157
’Image segmentation’ 165 219 161 196
’Vehicle’ 261 239 238 209
’ORL’ 230 200 264 228
’Colon Cancer’ 150 216 162 150
’Leukemia’ 140 240 140 176
These results show that the features selected by
C
4
provide accuracy rates that are higher than those
obtained by features selected by the classical scores.
This same conclusion was found in our earlier work
that follows the classical supervised evaluation of the
constraint scores (Kalakech et al., 2011) (Kalakech
et al., 2010).
4.4 Accuracy vs. Number of
Constraints
We compare the accuracy rates of the different scores
with a fixed number of features vs. number of pair-
wise constraints.
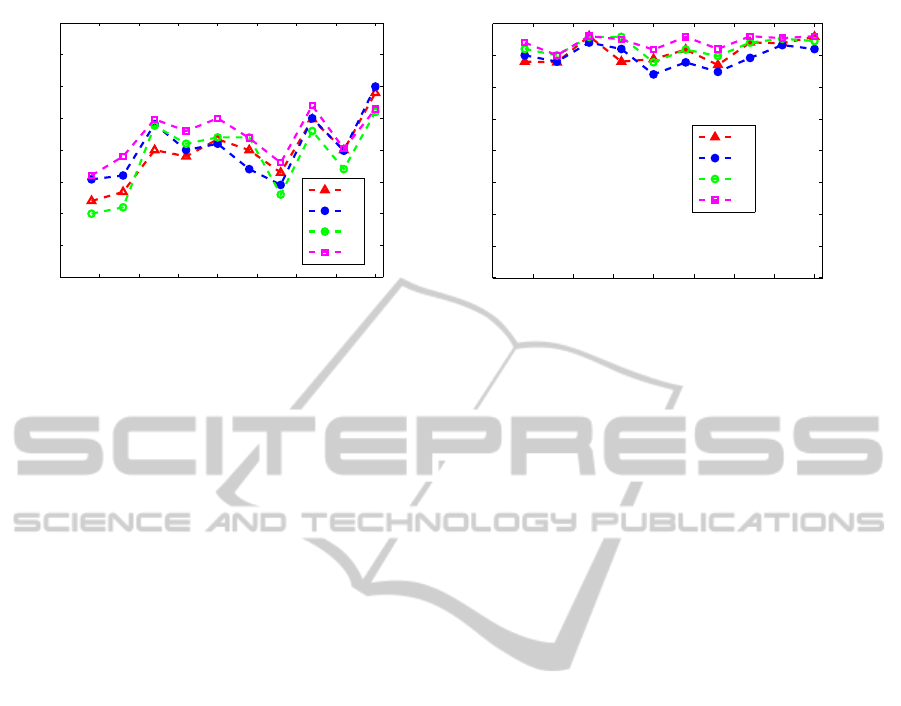
Figure 3 displays the plot of accuracy with a fixed
number of selected features (half of the number of
original features (Sun and Zhang, 2010) vs. differ-
ent number of pairwise constraints on the two gene
expression databases. The accuracy of the test sam-
ples is measured thanks to the supervised evaluation
scheme.
From this figure, we can see that, for almost all the
number of constraints, our score C
4
provides higher
accuracy than C
1
, C
2
and C
3
. We can also notice that
the accuracy does not tend to increase with respect
to the number of constraints. This is due to the con-
straints choice. Since these constraints are randomly
generated, some of them could be less informative
than the others.
Table 2: The rank sum of the constraint scores for different
number 2l of constraints on the ’Colon Cancer’ database.
2l\T T
1
T
2
T
3
T
4
4 constraints 140 157 153 224
10 constraints 242 147 201 153
40 constraints 153 268 169 124
60 constraints 257 171 194 151
Furthermore, Tables 2 and 3 show the rank sum
T
∗
for different number 2l of constraints (4, 10, 40
and 60) on the ’Colon Cancer’ and the ’Leukemia’
databases, respectively. The rank sum of each of the
semi-supervised criteria is calculated with the half of
the original features of each of the gene expression
Table 3: The rank sum of the constraint scores for different
number 2l of constraints on the ’Leukemia’ database.
2l\T T
1
T
2
T
3
T
4
4 constraints 194 215 210 190
10 constraints 234 176 159 150
40 constraints 148 198 196 123
60 constraints 167 170 184 115
databases.
We can see that, for the ’Colon Cancer’ database,
our score provides the lowest rank sum T (indicated
in bold) for 2 times over the 4 rows of Table 2 (when
the number of constraints is higher than 10). For the
’Leukemia database’, our score provides the lowest
rank sum T (indicated in bold) for the different num-
bers of constraints (4, 10, 40 and 60). These results
show that the features selected thanks to our score C
4
provide accuracy rates which are higher than those
obtained by the features selected by constraint scores
C
1
, C
2
and C
3
. The same conclusions were found
using the classical supervised evaluation (Kalakech
et al., 2011).
5 CONCLUSIONS
The accuracy rates of test samples reached by a clas-
sifier that analyses the features selected by the con-
straint scores, are generally compared in the super-
vised learning context. The nearest neighbor clas-
sifier uses the training sample labels as prototypes
of classes. However, the feature selection has been
performed in a semi-supervised context, since it uses
the available constraint sets and/or the unconstrained
samples.
So, we have proposed in this paper, to keep the
same learning context for the feature selection as for
the evaluation. The prior knowledge represented by
must-link and cannot-link constraints is used for the
selection and for the classification. The training sam-
ple labels are estimated by using the constrained K-
means algorithm that tries to respect the constraints
as much as possible. These estimated labels are then
used as prototypes by the nearest neighbor classifier
to classify the test samples. We called this approach
semi-supervised evaluation to distinguish it from the
classical supervised one.
The comparison between the different constraint
scores thanks to this semi-supervised evaluation
shows that the accuracy rates provided by our score
are higher than those of the classical scores.
We notice that the constrained K-means used dur-
ing the semi-supervised evaluation is a simple cluster-
ing algorithm. It does not guarantee the respect of all
SEMI-SUPERVISED EVALUATION OF CONSTRAINT SCORES FOR FEATURE SELECTION
181
0 5 10 15 20 25 30 35 40
0.3
0.35
0.4
0.45
0.5
0.55
0.6
0.65
Number of constraints
Accuracy
C
1
C
2
C
3
C
4
(a) ’Colon Cancer’.
0 5 10 15 20 25 30 35 40
0.3
0.35
0.4
0.45
0.5
0.55
0.6
0.65
Number of constraints
Accuracy
C
1
C
2
C
3
C
4
(b) ’Leukemia’.
Figure 3: Accuracy rates vs. number of constraints for C
1
, C
2
, C
3
and C
4
on the gene expression databases thanks to the
semi-supervised evaluation scheme. The desired number of selected features is half of the number of the original features.
the constraints specially when several constraints are
defined with the same sample. So, it will be interest-
ing to use another constrained classification algorithm
that is more efficient than this one, as these presented
by Kulis et al. (Kulis et al., 2009) and Davidson et al.
(Davidson et al., 2006).
REFERENCES
Alon, U., Barkai, N., Notterman, D., Gishdagger, K., Ybar-
radagger, S., Mackdagger, D., and Levine, A. (1999).
Broad patterns of gene expression revealed by cluster-
ing analysis of tumor and normal colon tissues probed
by oligonucleotide arrays. Proceedings of the Na-
tional Academy of Science of the USA, 96(12):745–
6750.
Blake, C., Keogh, E., and Merz, C. (1998). UCI
repository of machine learning databases.
http://www.ics.uci.edu/ mlearn/MLRepository.html.
Carpaneto, G. and Toth, P. (1980). Algorithm 548: solu-
tion of the assignment problem. ACM Transactions
on Mathematical Software.
Davidson, I., Wagstaff, K., and Basu, S. (2006). Measur-
ing constraint-set utility for partitional clustering al-
gorithms. In In proceedings of the Tenth European
Conference on Principles and Practice of Knowledge
Discovery in Databases ’PKDD06’, pages 115–126,
Berlin, Germany.
Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasen-
beek, M., Mesirov, J. P., Coller, H., Loh, M. L., Down-
ing, J. R., Caligiuri, M. A., and Bloomfield, C. D.
(1999). Molecular classification of cancer: class dis-
covery and class prediction by gene expression moni-
toring. Science, 286:531–537.
He, X., Cai, D., and Niyogi, P. (2005). Laplacian score
for feature selection. In Proceedings of the Advances
in Neural Information Processing Systems (’NIPS
05’), pages 507–514, Vancouver, British Columbia,
Canada.
Kalakech, M., Biela, P., Macaire, L., and Hamad, D. (2011).
Constraint scores for semi-supervised feature selec-
tion: A comparative study. Pattern Recognition Let-
ters, 32(5):656–665.
Kalakech, M., Porebski, A., Biela, P., Hamad, D., and
Macaire, L. (2010). Constraint score for semi-
supervised selection of color texture features. In Pro-
ceedings of the third IEEE International Conference
on Machine Vision (ICMV 2010), pages 275–279.
Kudo, M. and Sklansky, J. (2000). Comparison of algo-
rithms that select features for pattern classifiers. Pat-
tern Recognition, 33(1):25–4.
Kulis, B., Basu, S., Dhillon, I., and Mooney, R. (2009).
Semi-supervised graph clustering: a kernel approach.
Machine Learning, 74(1):1–22.
Liu, H. and Motoda, H. (1998). Feature extraction con-
struction and selection a data mining perspective.
Springer, first edition.
Samaria, F. and Hartert, A. (1994). Parameterisation of
a stochastic model for human face identification. In
Proceedings of the Second IEEE Workshop on Appli-
cations of Computer Vision ’ACV 94’, pages 138–142,
Sarasota, Florida.
Sun, D. and Zhang, D. (2010). Bagging constraint score
for feature selection with pairwise constraints. Pattern
Recognition, 43:2106–2118.
Wagstaff, K., Cardie, C., Rogers, S., and Schroedl, S.
(2001). Constrained k-means clustering with back-
ground knowledge. In Proceedings of the Eigh-
teenth International Conference on Machine Learning
’ICML 01’, pages 577–584, Williamstown, MA, USA.
Zhang, D., Chen, S., and Zhou, Z. (2008). Constraint score:
A new filter method for feature selection with pairwise
constraints. Pattern Recognition, (41):1440–1451.
Zhao, J., Lu, K., and He, X. (2008). Locality sensitive semi-
supervised feature selection. Neurocomputing, 71(10-
12):1842–1849.
Zhao, Z. and Liu, H. (2007). Semi-supervised feature selec-
tion via spectral analysis. In Proceedings of the SIAM
International Conference on Data Mining ’ICDM 07’,
pages 641–646, Minneapolis.
NCTA 2011 - International Conference on Neural Computation Theory and Applications
182
