
TRAINING RADIAL BASIS FUNCTION NETWORKS
BY GENETIC ALGORITHMS
Juliano F. da Mota
1,3
, Paulo H. Siqueira
2,3
, Luzia V. de Souza
2,3
and Adriano Vitor
1,3
1
Department of Mathematics, Paraná State University, Com. Norberto Marcondes Avenue 733, Campo Mourão, Brazil
2
Department of Graphical Expression, Paraná Federal University, Cel Francisco H. dos Santos Avenue, Curitiba, Brazil
3
Graduate Program in Numerical Methods in Engineering, Paraná Federal University, Curitiba, Brazil
Keywords:
Radial basis function neural networks, Evolutionary computation, Pattern classification.
Abstract:
One of the issues of modeling a RBFNN - Radial Basis Function Neural Network consists of determining the
weights of the output layer, usually represented by a rectangular matrix. The inconvenient characteristic at
this stage it’s the calculation of the pseudo-inverse of the activation values matrix. This operation may become
computationally expensive and cause rounding errors when the amount of variables is large or the activation
values form an ill-conditioned matrix so that the model can misclassify the patterns. In our research, Genetic
Algorithms for continuous variables determines the weights of the output layer of a RBNN and we’ve made
a comparsion with the traditional method of pseudo-inversion. The proposed approach generates matrices of
random normally distributed weights which are individuals of the population and applies the Michalewicz’s
genetic operators until some stopping criteria is reached. We’ve tested four classification patterns databases
and an overall mean accuracy lies in the range 91–98%, in the best case and 58–63%, in the worse case.
1 INTRODUCTION
For a long time scientists have been trying to develop
methods for pattern classification problems wich may
help a decision taker in a uncertain scenario. Several
mathematical and statistical methods had been devel-
oped and, for this reason, improvements in existents
methods and the building up of new methods wich
may reduce the computational effort, offer better re-
sults or both are required nowadays.
Every single existent method offer a different op-
tion relative to speed and quality of the prediction or
classification. The most difficult task, wich every re-
searcher yearns, is to develop a method capable of get
a high accuracy in a minimal time given the need for
speed of the on-line era. By “high accuracy” we mean
an error as small as possible and a correct classified
percentual as big as possible, considering a pattern
classification problem.
This study compares two training methods of a
RBFNN, one of them is considered traditional, which
is based in calculating the pseudo-inversion of a rect-
angular matrix and the other uses genetic algorithms
for continuous data, changing this matrix into a ge-
netic population individual, and the main objective is
to find the optimal (or near-optimal) matrix through
natural selection and genetic operators.
To reach this goal, the basic precepts of Radial
Basis Function Neural Networks and Genetic Algo-
rithms are presented and its main features are ex-
plained in order to form a conceptual basis for the
experiments.
2 SOME RELATED
RESEARCHES
Trying to solve the main problem of the classical ap-
proach of training a RBFNN, which is the need to
calculate the pseudoinverse of a rectangular matrix,
some authors have proposed alternative methodolo-
gies to change that problem into another of lower
computational complexity.
The researchers (Li and Ling, 2011) applied the
idea of getting the weights of the hidden layer of a
RBFNN to develop a generalized model predictive
control in a power generating unit that had two in-
put and two output variables. The experiment was to
compare the performance of the proposed algorithm
(called GA-RBF) with the performance of a Multi-
layer Perceptron using the back-propagation training
373
F. da Mota J., H. Siqueira P., V. de Souza L. and Vitor A..
TRAINING RADIAL BASIS FUNCTION NETWORKS BY GENETIC ALGORITHMS.
DOI: 10.5220/0003751903730379
In Proceedings of the 4th International Conference on Agents and Artificial Intelligence (ICAART-2012), pages 373-379
ISBN: 978-989-8425-95-9
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

algorithm. The proposed technique has a mean square
error of the order of 10
−5
, while the Perceptron tested
missed about 10
−1
.
The research of (Changbing and Wei, 2010) ap-
plied RBFNN trained by GA to forecast the risk in
an aquatic environment. The tests show that the pro-
posed model outperformed the "gray model" which
uses a first order differential equation to estimate val-
ues based on historical data. There are charts on
which you can visualize the superiority of the pro-
posed model to predict the risk.
By the other hand (Ming et al., 2010) applied the
same basic idea for predicting the flow in a network.
The results of the experiment, in which the flow of
a network with 40 points was predicted, show that
the relative error in the prediction of a RBFNN train-
ing via the pseudo-inversion (traditional) was 0.0247
while the RBFNN trained by GA could miss only
0.0177, a reduction of almost 30% in error.
The research (Kurban and Be¸sdok, 2009) com-
pared four algorithms to train a RBFNN: Bee Colony,
GA, Kalman filter and gradient descent. They tested
three databases available in (Frank and Asuncion,
2010) and one database application proposed by the
authors. In all of tested datasets the Bee Cllony algo-
rithm performances was slightly higher than the GA,
both featuring an accuracy above 90% accuracy the
test set in the database in (Frank and Asuncion, 2010)
and over 70% of the database application in sensors
proposed by the authors.
3 RADIAL BASIS FUNCTION
NEURAL NETWORKS
A RBFNN is feed-forward and it has only two layers,
one of them is a hidden layer and the other is the out-
put layer. In the hidden layer the activation functions
of neurons are radial basis functions. A RBF - Ra-
dial Basis Function is defined by (Haykin, 2001) as
any function that its functional values are equal to the
norm of its arguments.
3.1 Neural Networks wich uses Radial
Basis Functions
The learning process of this network has its founda-
tions in the theory of nonlinear programming. The ar-
chitecture of a RBFNN is quite simple, there are only
two layers in addition to the input nodes, one hidden
layer, which has radial basis functions as activation
functions and an output layer which has linear func-
tions as activation, as seen in Figure 1.
x
1
c
1
c
2
c
s
.
.
.
.
.
.
w
11
w
21
w
p1
w
12
w
22
w
p2
w
1s
w
2s
w
ps
φ
1
φ
2
φ
s
y
1
y
2
y
p
w
10
w
20
w
p0
1
1
1
Figure 1: Representation of the Architecture of a RBFNN.
Each neuron in the hidden layer has an associated
vector, called the center of the neuron, which defines
the center of the receptive field of that neuron. Gen-
erally such vectors are stored in a C matrix, called the
matrix of centers of neurons. These vectors have a
strong influence on network performance. Later, in
section 3.2 some methods will be briefly presented.
The m-th activation value from a hidden layer neu-
ron depends of the distance between the input node x
i
and the center c
j
, where c
j
∈ C is the center of the
m-th neuron. Thus, by the classical approach, train a
RBFNN is equivalent to calculate the wheights matrix
w to fit each y to one target t, as shows the equation 1,
y
r
= w
0r
φ
0r
+
s
∑
k=1
w
kr
φ
||x
i
−C||
. (1)
3.2 Neurons Centers Selection
The task of selecting the neurons centers is essentially
a clustering problem. There are a few classic strate-
gies described in (Haykin, 2001) and a brief discus-
sion of these strategies is presented below.
3.2.1 Random Fixed Centers
This is the simplest and least expensive way to se-
lect the centers. Although simple, it is considered by
(Haykin, 2001) as “the most sensible approach” be-
cause in each experiment a different part of the ma-
trix of observations is used, and for this reason, the C
matrix will contain a good representation of the data
space.
The limitation of this method lies in the need of
a large training set to achieve a satisfactory perfor-
mance. Possibly, the standard most commonly used
training 60–20–20, meaning 60% for the training set
and 20% validation and testing, would probably have
to be modified and hence the model could lose the
ability to generalize.
ICAART 2012 - International Conference on Agents and Artificial Intelligence
374
3.2.2 Self-organized Selection of Centers
Also described in (Haykin, 2001) and (Silva et al.,
2010), this method consists of the main idea of the
SOM - Self Organizing Map Algorithm proposed by
Kohonen apud (Haykin, 2001), wich is described be-
low:
1. Choose distinct random values as initial centers,
in most cases elements of the training set;
2. During the n-th iteration, take a sample of training
set;
3. Find the center with the minimal euclidian dis-
tance to each entering vector;
4. Update the winner center position acording the
equation:
c(new) = c(old) + η(x− c) (2)
where η ∈ (0, 1) is a learning rate, x is the entering
vector and c is the winner center.
5. Back to Step 2 until no noticiable changes in the
centers matrix can be perceived.
The main difference between this algorithm and
the classical SOM algorithm is precisely the absence
of a neurons map, i.e., no neighborhood is consid-
ered to update the neurons. That means to say a SOM
with the "Winner Takes All" principle (Siqueira et al.,
2005). where only the winner neuron is updated.
3.2.3 Supervisioned Selection of Centers
This is the most generic form to make the selection of
centers. The idea is to fit the centers by error correc-
tion and thus RBFNN resembles the classical Percep-
tron.
4 CONTINUOUS DATA GENETIC
ALGORITHMS
The GA - Genetic Algorithm as described in (Hol-
land, 1975) apud (Man et al., 1996) are basically a set
of algorithms based on the principles of evolutionary
biology stated by Charles Darwin in his book known
worldwide The Evolution of Species. The main objec-
tive of the GAs is to optimize functions.
The considered principles in a GA preparation are
natural selection, heredity, mutation and recombina-
tion (crossing-over). When translating into a math-
ematical language, these principles become genetic
operators of crossover and mutation, such a way that
heritability and natural selection becomes a decision
rule.
The basic algorithm, systematized by (Holland,
1975) apud (Man et al., 1996), has low implemen-
tation complexity and this is one of the great advan-
tages of this technique. The steps of the algorithm are
described in the algorithm in Figure 2.
It is noteworthy that, initially, the binary number
base (binary representation) is considered for imple-
mentation and application of GAs. Today, however,
is quite common to find applications where the dec-
imal number base is used. Possibly, this change has
been due to real representation allow greater range of
operators, as will be shown in section 4.1.
Let g = 0 the generation counter;
Create and initialize a presized population;
while no stop criteria is reached do;
Evaluate the fitness of each individual in population;
Perform reproduction to generate offspring;
Select the new population;
Advance to the new generation, i. e., g = g+ 1;
end while
Figure 2: The GA Algorithm.
4.1 Real Representation and
Michalewicz Operators
One of the studies using the real representation can
be found in (Michalewicz et al., 1994), were there
are three crossover operators and three mutation op-
erators described and mathematically justified. Ad-
ditionally to those descriptions, we describe also an
extra mutation operator. In all operators, p
j
are the
parents and c
j
are the offspring.
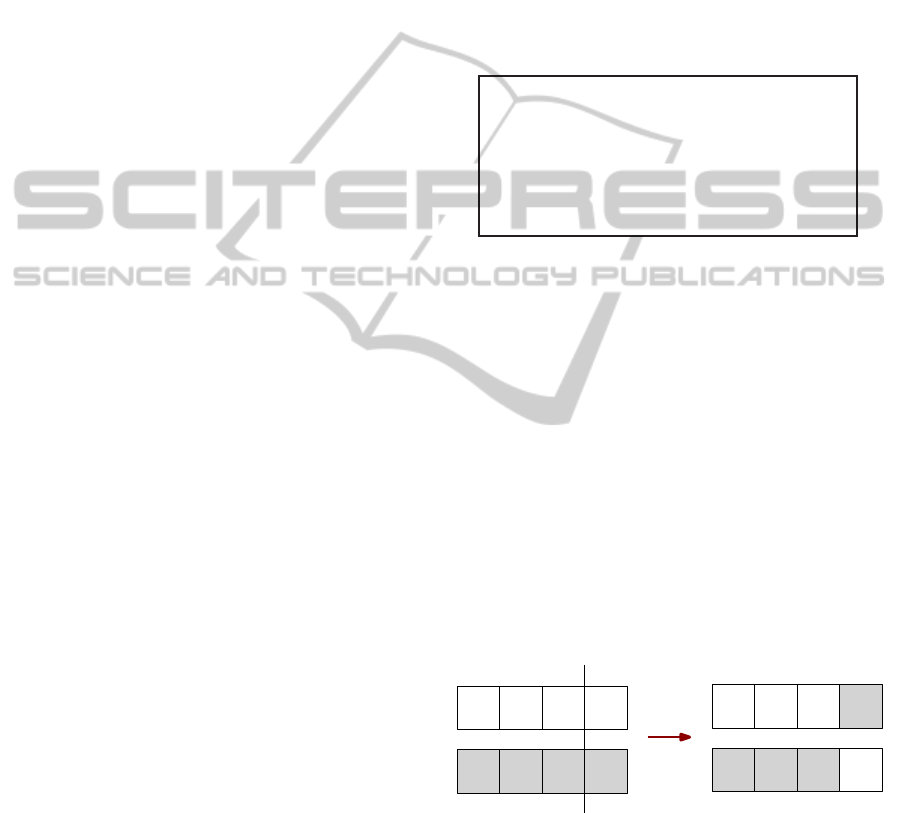
The ordinary crossover is a variation of a conven-
tional one point crossover, which is used in the binary
representation, adapted to the real representation, as
we can see in Figure 3.
1,5 9,2
31,2
6,8
0,8
7,4
1,3
5,7
1,5
9,2
31,2
6,80,8
7,4
1,3
5,7
Parents Offspring
Figure 3: One Point Crossover Representation.
Given two individuals, the arithmetical crossover
generates,
c
1
= βp
1
+ (1− β)p
2
and
c
2
= βp
2
+ (1− β)p
1
with β ∼ U(0, 1).
For another two individuals such that the fitness of p
1
is greater than the fitness of p
2
, the heuristic crossover
TRAINING RADIAL BASIS FUNCTION NETWORKS BY GENETIC ALGORITHMS
375

generates,
c = p
1
+ β(p
2
− p
1
) where β ∼ U(0, 1).
Given an individual p, the uniform mutation oper-
ator replaces one gene by a random number coming
from a uniform distribution, i.e.,
c
i
=
U(a
i
, b
i
), if i = j
p
i
, otherwise.
The values a
i
and b
i
represents the interval limits
to the individual c
i
, just in case of a factibility restric-
tion.
Given an individual p, the limit mutation operator
replaces one gene by one of the interval limits [a
i
, b
i
],
avoiding the arithmetical crossover to take the genes
to the interval center, here we consider r ∼ U(0, 1),
c
i
=
a
i
, if r < 0, 5 and i = j
b
i
, if r > 0, 5 e i = j
c
i
, otherwise .
Finally, an individual p, the non-uniform mutation
operator replaces one gene by a random number com-
ing from a non-uniform distribution, i.e.,
c
i
=
p
i
+ (b
i
− p
i
) f(G), if r
1
< 0, 5 and i = j
p
i
− (p
i
− a
i
) f(G), if r
1
≥ 0, 5 and i = j
p
i
, otherwise .
considering,
f(G) =
r
2
1−
G
G
max
b
,
where G is the current generation, G
max
is the maxi-
mum number of generations and r
1
, r
2
∼ U(0, 1). The
application of non-uniform mutation in all genes of
this individual is called multiple non-uniform muta-
tion.
5 EXPERIMENTS AND RESULTS
ANALYSIS
In our experiments, the performance of a RBFNN us-
ing the traditional method of training, the pseudo-
inversion of the matrix that contains the activa-
tion values of the intermediate layer, was com-
pared with alternative training pathway GA, con-
sidering Michalewicz operators (Michalewicz et al.,
1994). We’ve used the already well-known classifi-
cation problems - Iris, Contraceptive Method Choice
(CMC), Cancer and Blood Transfusion (BT) - data
sets that are found in (Frank and Asuncion, 2010).
The characteristics of the sets used in the tests are
Table 1: Datasets Features.
Dataset Entries Outputs Training Validation Test
Iris 4 3 90 30 30
CMC 9 3 884 295 294
Cancer 9 2 420 140 139
BT 4 2 449 150 149
shown in Table 1, the number of variables in standard
input, output and the number of patterns in each set.
For all data sets, we performed an experiment that
consisted of five steps:
1. Separate the database into three sets:
• Training set, with 60% of the examples;
• Validation set, with 20% of the examples and;
• Set of tests, with 20% of the examples.
2. Train a RBFNN 100 epochs with each approach
(pseudo-inversion and GA) and obtain CCP - Cor-
rect Classified Percentage in each set of item 1,
especially set of tests;
3. Record the results of the statistical percentages of
correct classifications of the two training meth-
ods;
4. Considering a significance level of 5% to test the
hypothesis of equality of variances in order to
determine which test comparing the means used,
writing H = 0 if variances are equal and H = 1
otherwise;
5. Considering a significance level of 5%, perform
two comparison tests medium with the following
assumptions:
First Test Second Test
h
0
: Average PI = Average GA h
0
: Average PI = Average GA
h
1
: Average PI > Average GA h
2
: Average PI < Average GA
considering h
1
= 0 if h
0
is not rejected in the first test
and h
1
= 1, oterwise. Considering yet h
2
= 0 if h
0
is
not rejected in the second test and h
2
= 1, otherwise.
According to the literature, one of the most im-
portant parameters for a neural network, including the
RBFNN, is the number of neurons in its layers, in the
specific case of this research, the amount of interme-
diate layer neurons. The limit here is set to test from
two to 10 neurons, increasing of two at each experi-
ment.
Two other major issues, when talking about the
parameters of a RBFNN, are the method of centers se-
lection and the definition of the spread of the Gaussian
activation functions in the hidden layer. The method
of centers selection used in this study was the self-
organized selection of centers, described in Section
3.2.2 and the method for setting the spread of each
RBF is described in (Silva et al., 2010) and basically
ICAART 2012 - International Conference on Agents and Artificial Intelligence
376

consists into calculating the radius of each function
based on the mean square distance between the inputs
and the centers of the neurons that received an update
during the self-organized selection of centers.
The parameters selection of a GA also does not
have a consolidated method, varying according to
each application. Therefore, it is necessary to perform
some initial tests to observe the influence of each pa-
rameter in the performance of the algorithm. In this
research, we’ve made more than 50 preliminary tests
and the best parameter setting is in Table 2.
Table 2: GA Parameters.
Parameter Value
Initial Population 100
Probability Crossing Operators 80%
Probability Mutation Operators 10%
Maximum Number of Generations 100
Talking about the mutation operators wich have
restricted the range to the generated numbers, i. e.,
the Limit Mutation, Non-uniform Mutation and Non-
uniform Multiple Mutation, the range adopted in all
cases was to accept an increase of up to 100% of the
highest absolute value of a gene in any individual.
This means that if the highest absolute value
among the genes of all individuals is τ, then the possi-
ble range for the generation of a mutation gene would
be (−2 · τ, 2 · τ). This measure sought a balance be-
tween excessiveextrapolation/restriction of the search
space.
In addition, with respect to the size of the initial
population we tested several initial population sizes
and the best result was obtained by the size of 100.
Although Genetic Algorithms are known for provid-
ing approximate solutions, here it’s being used as a
optimal solution pursuing technique.
The statistical results of 100 rounds for each data
set are in the Table 3–6, for Iris, CMC, Cancer and
BT, respectively. In the tables you can see informa-
tion about the mean and standard deviation of the per-
formance of each algorithm for each data set. In the
tables, the variable “NNeuro” represents the number
of neurons in the hidden layer. For each number of
neurons in the first line depicts the statistics using
pseudo-inversion and the second statistics by the GA
approach.
As shown in Table 3, the proposed model using
GA to obtain the weight matrix showed a slightly
greater variability than the approach via pseudo-
inversion. The mean CCP by the GA approach was
not superior to the pseudo-inversion in any case and
according to the test t, for comparison between two
means, there was a tie with two neurons and superi-
ority of the approach via pseudo-inversion with other
amounts of neurons tested, notice that in all tests the
considered significance level is 5 %.
Table 3: Results of Iris Set.
NNeuro Average±Std Dev Min Max H h
1
h
2
2
90, 5%± 5, 6% 73,3% 100%
1 0 0
88, 4%± 7, 8% 63,3% 100%
4
92, 3%± 5% 73,3% 100%
1 1 0
87, 3%± 7% 66,7% 96,7%
6
93, 9%± 3, 9% 86,7% 100%
1 1 0
88, 7%± 5, 6% 73,3% 100%
8
94, 6%± 3, 8% 83,3% 100%
1 1 0
87, 1%± 7, 3% 70% 100%
10
94, 6%± 3, 8% 86,7% 100%
1 1 0
85, 2%± 8, 1% 63,3% 100%
The results in Table 4 shows that considering a
significance level of 5%, the variance of the perfor-
mances of both techniques is not statistically sig-
nificant and there is superiority of the approach via
pseudo-inversion in two cases: six and 10 neurons in
the hidden layer, given the values of h
1
. Even compar-
ing the best performance in both cases, the approach
via GA was inferior about two percentage points ap-
proximately.
Table 4: Results of Contraceptive Method Choice Set.
NNeuro Average±Std Dev Min Max H h
1
h
2
2
57%± 2, 7% 49% 63,3%
0 0 0
56, 7%± 2, 9% 49% 63,3%
4
57, 1%± 2, 2% 52,7% 61,2%
0 0 0
56, 7%± 2, 6% 50,7% 62,6%
6
58, 5%± 3, 3% 51,7% 67,7%
0 1 0
56, 9%± 3, 2% 49,7% 64,3%
8
58, 2%± 3, 2% 50,7% 65,6%
0 0 0
57, 1%± 3, 2% 48,6% 62,9%
10
60, 5%± 2, 8% 52% 65%
0 1 0
58, 4%± 2, 7% 51,4% 64%
For the Cancer dataset in Table 5, considering a
significance level of 5%, there was no difference be-
tween the performance variances of both techniques,
except with 10 neurons, and that the only case in
which there was a statistically significant difference,
with six neurons in the hidden layer, this case was fa-
vorable to the GA approach given the h
2
value.
Finally, in Table 6 considering 5% of significance,
there was no difference between the variances of the
performance and the pseudo-inversion approach was
better in three of five numbers of neurons tested: six,
eight and 10 neurons. Once the volume of two four
neurons there was a tie.
TRAINING RADIAL BASIS FUNCTION NETWORKS BY GENETIC ALGORITHMS
377

Table 5: Results for the Cancer Dataset.
NNeuro Average± Std Dev Min Max H h
1
h
2
2
95, 6%± 1, 5% 92,1% 98,6%
0 0 0
96%± 1, 5% 92,9% 98,6%
4
95, 3%± 1, 7% 92,1% 98,6%
0 0 0
95, 9%± 1, 4% 92,9% 98,6%
6
95, 1%± 1, 7% 90,7% 97,9%
0 0 1
96%± 1, 6% 92,1% 98,6%
8
96%± 1, 2% 94,3% 98,6%
0 0 0
96, 3%± 1, 3% 93,6% 98,6%
10
95, 8%± 1, 6% 92,1% 100%
1 0 0
96, 2%± 1, 4% 93,6% 99,3%
Table 6: Results for Blood Transfusion Dataset.
NNeuro Average± Std Dev Min Max H h
1
h
2
2
75, 9%± 3, 2% 68,5% 83,9%
0 0 0
75, 9%± 3, 2% 68,5% 83,9%
4
77, 7%± 3, 3% 70,5% 84,6%
0 0 0
77, 2%± 3, 2% 71,1% 85,2%
6
78, 8%± 3, 9% 67,8% 89,3%
0 1 0
77, 5%± 3, 4% 67,8% 85,2%
8
77, 5%± 2, 4% 71,1% 81,9%
0 1 0
75, 9%± 3% 67,8% 81,9%
10
77, 6%± 3, 5% 71,8% 85,2%
0 1 0
76%± 3, 5% 68,5% 84,6%
6 CONSIDERATIONS AND
FUTURE WORK
Considering the results obtained and previously dis-
cussed, we can say that:
• the genetic operators presented did not produced
good populations, given that the GA approach did
not overcome the traditional approach, except in
one case that can be seen in Table 5;
• the GA approach performance did not reach the
expectations when compared with the traditional
approach, because even in the situation was better,
this superiority was not unquestionable;
• there were two cases where none of the two ap-
proaches achieved a significant result: in the data
set “Contraceptive Method Choice”, the best per-
formances of both algorithms was below 70% on
all executions and below 60% on average. In the
case of the “Blood Transfusion”, despite the mean
CCP was higher 70%, a possible application in
reality would not be feasible because the model
would not be reliable;
• the training via GA produced results lower than
the pseudo-inversion in three of four sets tested,
considering the significance defined in the tests.
Taking an overview of the results, considering a
5% level of significance, we can say that the GA ap-
proach even being able to produce results as good
as the supposedly exact approach had a significantly
lower performance in the databases we’ve tested.
However, it is necessary to consider that the exact ap-
proach suffers with complexity and numerical round-
ing errors with the growth of the number of neurons in
the hidden layer. Thus, taking into account the adapt-
ability of GA, if new more robust genetic operators
are developed will overcome this limitation.
6.1 Future Works
Noting the apparent failure of the considerations
made in this study, below we’ve listed some possi-
ble suggestions for future works that may continue or
refute this line of thinking:
• test other GA operators;
• compare other search algorithms (and GA) with
the pseudo-inversion approach;
• compare other search algorithms with supervised
learning approach;
• use statistical techniques such as Principal Com-
ponent Analysis and Fator Analysis in the treat-
ment of data in order to see if they can help im-
prove performance in sets with similar character-
istics o the "Contraceptive Method Choice ", in
which both approaches had less than 70% perfor-
mance.
REFERENCES
Changbing, L. and Wei, H. (2010). Application of genetic
algorithm-rbf neural network in water environment
risk prediction. 2nd International Conference on
Computer Engineering and Technology 2010, pages
239–242.
Frank, A. and Asuncion, A. (2010). Uci machine learning
repository.
Haykin, S. (2001). Redes neurais - princípios e práticas.
Bookman.
Holland, J. (1975). Adaption in Natural and Artificial Sys-
tems. MIT Press.
Kurban, T. and Be¸sdok, E. (2009). A comparsion of rbf
neural netowrk training algorithms for inertial sen-
sor based terrian classification. Sensors, pages 6312–
6329.
Li, N. and Ling, H. (2011). Study of an algorithm of ga-
rbf neural network generalized predictive control for
generating unit. International Conference on Elec-
tric Information and Control Engineering 2011, pages
1723–1726.
Man, K., Tang, K., and Kwong, S. (1996). Genetic algo-
rithms: concepts and applications. IEEE Transactions
on Industrial Electronics, 43(5):519–534.
ICAART 2012 - International Conference on Agents and Artificial Intelligence
378

Michalewicz, Z., Logan, T. D., and Swaminathan, S.
(1994). Evolutionary operators for continuous con-
vex parameter spaces. Proceedings of the 3rd Annual
Conference on Evolutionary Programming, pages 84–
97.
Ming, Z. Y., Bin, Z. Y., and Zhong, L. L. (2010). Appli-
cation of genetic algorithm and rbf neural network in
network flow prediction. 3rd IEEE International Con-
ference on Computer Science and Information Tech-
nology 2010, pages 298–301.
Silva, I. N., Spatti, D. H., and Flauzino, R. A. (2010). Redes
neurais artificiais - para engenharia e ciências apli-
cadas. Artliber.
Siqueira, P., Scheer, S., and Steiner, M. T. A. (2005). Ap-
plication of the "winner takes all" principle in wang’s
recurrent neural network for the assignment problem.
Lecture Notes in Computer Science, 3496(1):731–
738.
TRAINING RADIAL BASIS FUNCTION NETWORKS BY GENETIC ALGORITHMS
379
