
ABSTRACTIONS FOR SCALING ESCIENCE APPLICATIONS TO
DISTRIBUTED COMPUTING ENVIRONMENTS
A StratUm Integration Case Study in Molecular Systems Biology
Per-Olov
¨
Ostberg
1
, Andreas Hellander
2,3
, Brian Drawert
3
,
Erik Elmroth
1
, Sverker Holmgren
2
and Linda Petzold
3
1
Dept. of Computing Science, Ume
˚
a University, SE-901 87, Ume
˚
a, Sweden
2
Uppsala University, SE-751 05 Uppsala, Sweden
3
University of California, Santa Barbara, CA 93106-5070 Santa Barbara, U.S.A.
Keywords:
Systems biology, eScience, Grid computing, Cloud computing, Service-oriented architecture.
Abstract:
Management of eScience computations and resulting data in distributed computing environments is compli-
cated and often introduces considerable overhead. In this work we address a lack of integration tools that
provide the abstraction levels, performance, and usability required to facilitate migration of eScience appli-
cations to distributed computing environments. In particular, we explore an approach to raising abstraction
levels based on separation of computation design from computation management and present StratUm, a com-
putation enactment tool for distributed computing environments. Results are illustrated in a case study of
integration of a software from the systems biology community with a grid computation management system.
1 INTRODUCTION
In this work we explore an approach to migrating
computational applications to virtual distributed com-
putational infrastructures based on separation of com-
putation design and enactment of computations in dis-
tributed computing environments. In a case study, we
investigate raising abstraction levels for distributed
eScience
1
computation management and integrate a
public-domain eScience application from the compu-
tational systems biology community with a frame-
work for management of scientific computations in
heterogeneous grid environments.
Current methodology for scaling computational
capacity beyond the capabilities of individual re-
source sites includes techniques such as aggregation
and federation of distributed resource systems (Grid
computing), and virtualization of resource sets for
provisioning of compute capacity as metered services
(Infrastructure-as-a-Service Cloud computing). Typ-
1
We define eScience applications to be computational
science applications operating in distributed computing,
e.g., grid computing, environments. In this work we focus
on the particular use cases of applications from the systems
biology field, but the techniques and tools developed are ap-
plicable to most forms of distributed scientific computing.
ically, the volatility and heterogeneity of such (Grid
and Cloud computing) resource sets make utilization
of fine-grained synchronization in computations in-
feasible. Instead, data and task parallelism are of-
ten exploited through organization of computations
as large numbers of autonomous tasks that can be
processed individually. The size and complexity of
eScience applications make the coordination of such
computations non-trivial. In addition, distributed vir-
tual environments typically also introduce substantial
complexity in the management of computations and
resulting data sets. Factors such as these necessitate
the use of abstractive high-level tools for computation
management in virtual computing infrastructures.
In a case study we focus on integration of a public
domain simulation software (URDME, Section 3.1)
and a grid computation management tool (the Grid
Job Management Framework (GJMF), Section 3.2).
As part of this effort, we have developed a computa-
tion management and integration architecture called
the Stratified Resource Abstraction Toolkit (StratUm,
Section 3.3), which is designed to separate compu-
tation design from computation management, raise
abstraction levels for computation management, and
provide versatility in computation enactment.
The resulting system demonstrates a viable design
290
Östberg P., Hellander A., Drawert B., Elmroth E., Holmgren S. and Petzold L..
ABSTRACTIONS FOR SCALING eSCIENCE APPLICATIONS TO DISTRIBUTED COMPUTING ENVIRONMENTS - A StratUm Integration Case Study
in Molecular Systems Biology.
DOI: 10.5220/0003765002900294
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2012), pages 290-294
ISBN: 978-989-8425-90-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

pattern for separation of eScience computation design
from infrastructure computation enactment, and illus-
trates how abstraction levels for computation man-
agement can be raised. The rest of this paper is or-
ganized as follows. Section 2 provides a brief back-
ground to the case study application, Section 3 gives
an overview of the integration project, and Section 4
discusses the resulting architecture. Section 5 sam-
ples related work, and Section 6 concludes the paper.
2 MESOSCOPIC SPATIAL
STOCHASTIC SIMULATION
An important theoretical tool for investigating the
properties of cellular regulatory systems is the con-
struction and simulation of quantitative models of
their dynamic behavior. In molecular systems biol-
ogy such models are frequently used with the aim to
gain a system-level understanding of basic regulatory
mechanisms that arise from experimentally known or
assumed macromolecule (e.g. protein) interactions.
A popular and widely used modeling framework
that accounts for stochasticity is the Markov process.
Simulated trajectories of models based on the Monte
Carlo methodology are typically independent, thus
the overall problem is inherently task-parallel and
maps well to distributed resource utilization patterns.
This characteristic is a feature shared among many
traditional eScience applications.
In the following sections we demonstrate an ap-
proach to scaling computations and data manage-
ment for a spatial stochastic simulation software
(URDME) to distributed computing resources using
an abstractive computation management architecture
(StratUm). The focus of this case study is directed
towards integration with computational grid environ-
ments through use of a middleware-agnostic grid job
management tool (GJMF),
3 DESIGN AND MANAGEMENT
OF COMPUTATIONS
Computations in eScience environments require large
amounts of computational power and data storage ca-
pacity. Efficient scaling of computations is compli-
cated by the complexity of computation management
in virtual computational infrastructures. Any suitable
design pattern that facilitates integration and extensi-
bility of eScience applications with distributed com-
putational resources will inevitably involve high-level
abstractions on both the job management level as well
as on the level of the computational software; core
simulation routines need to be functionally and struc-
turally decoupled from client user interfaces. Simi-
larly, resource access needs to be decoupled from the
computational application as well as from resource
site-specific software layers.
In this section we present results from a case study
integration of two systems: URDME, a public do-
main software package for spatial stochastic simula-
tion, and GJMF, a middleware-agnostic grid job man-
agement framework. In addition, we also present a
recently developed integration architecture, StratUm,
designed to abstract and reduce integration complex-
ity for both applications and infrastructures.
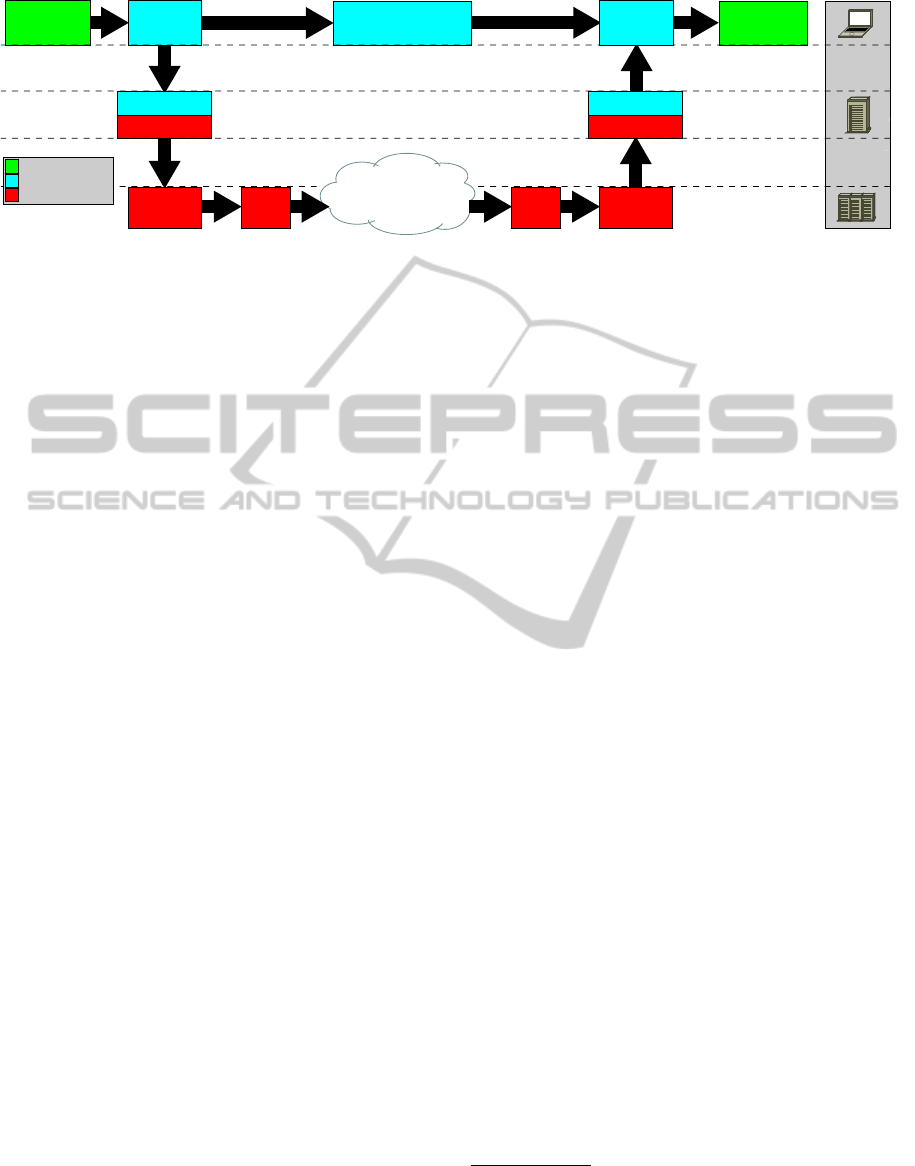
3.1 URDME
URDME is a software framework for spatial stochas-
tic simulation using unstructured meshes (Drawert
et al., 2011). It is designed to be a versatile tool
for both applied users and developers of new spa-
tial stochastic simulation algorithms. The top layer in
Fig. 1 shows an overview of the design of the package.
URDME relies on third party software for geometry
modeling, mesh generation and pre- and postprocess-
ing (green). A Matlab interface provides a familiar,
interactive, and flexible environment for model devel-
opment and provides a bridge to the core simulation
routines (cyan). Stochastic simulation algorithms are
implemented as stand-alone C/C++ executables.
As a part of the design pattern for the overall sys-
tem we have developed a new server-side component
to the URDME framework. In our implementation,
the desktop software is extended to include non-local
computation by implementing a URDME-server soft-
ware package (Fig. 1, middle layer) that is designed
to provide a transparent interface between the desktop
client and remote job management systems.
The overall architecture of this system has the
URDME client running on the user’s desktop, the
URDME-server running on a standalone server, the
StratUm and GJMF services running on separate
servers, and computations running on dedicated com-
putational resources in a distributed grid environment.
In this case study, the computational resources of
the Swedish national grid, SweGrid
2
, are accessed
through StratUm. The URDME-server communicates
with StratUm via native StratUm client APIs (Fig.
1, red). From the perspective of URDME, StratUm
provides a high-level, low-complexity interface to the
Grid Job Management Framework, GJMF (
¨
Ostberg
and Elmroth, 2010).
2
SweGrid: http://www.snic.vr.se/projects/swegrid
ABSTRACTIONS FOR SCALING eSCIENCE APPLICATIONS TO DISTRIBUTED COMPUTING ENVIRONMENTS
- A StratUm Integration Case Study in Molecular Systems Biology
291
3D Modeling
software
(Comsol)
URDME
3rd Party Software
StratUm Software
URDME Software
(Matlab)
Stochastic Solver
(C/C++)
Visualization /
Post-processing
(Matlab/Comsol)
URDME Server
StratUm Client
StratUm
Server
URDME Server
StratUm Client
StratUm
Server
SweGrid
Urdme
protocol
StratUm
protocol
URDME
(Matlab)
GJMF
GJMF
Figure 1: System process flow. The top layer shows the previous, client-only interactive workflow of URDME. The new
server-side component (middle layer) interacts with the StratUm server (bottom layer) through the StratUm API.
3.2 The Grid Job Management
Framework (GJMF)
The Grid Job Management Framework
(GJMF) (
¨
Ostberg and Elmroth, 2010) is a framework
for computation enactment in grid environments. The
GJMF is constructed as a hierarchically ordered set
of services, where higher level services aggregate
the capabilities of lower level services. Services are
connected using dynamic configuration (composi-
tion) techniques. The framework offers a unified set
of job management interfaces that provides access
to multiple grid middleware concurrently. The
architecture of the GJMF organizes services in layers
that offer increasingly advanced job management
functionality. Services in higher layers aggregate the
functionality of services in lower layers and offer
higher abstraction levels and automation, while lower
layer services provide more fine-grained job control.
The GJMF architecture is constructed as a net-
work of services, which provides an architecture
model where services are dynamically composed, and
includes dynamic fail-over capabilities through re-
dundancy. Services can function as individual stan-
dalone services and framework components simul-
taneously. The architectural model of the GJMF
provides great flexibility in system deployment, the
framework itself can for example be dynamically re-
configured during runtime. The GJMF is imple-
mented in Java, is constructed using the Globus
Toolkit 4 (GT4) (Foster, 2005), and utilizes Web Ser-
vice Resource Framework (WSRF) notifications for
state coordination.
3.3 The Stratified Resource Abstraction
Toolkit (StratUm)
The GJMF provides a generic and flexible framework
for computation enactment in federated grid environ-
ments. As the GJMF builds on the Globus Toolkit and
utilizes WSRF notifications, integration of computa-
tion tools like URDME and the GJMF can sometimes
be complex tasks. To reduce the integration footprint
and facilitate more flexible utilization of the GJMF
for scientific computations, we have developed an
extended architecture integration model dubbed the
Stratified Resource Abstraction Toolkit (StratUm
3
).
Here, StratUm functions as an integration bridge be-
tween URDME and GJMF, and is used to explore
integration models and functionality abstractions in
eScience environments. As illustrated in Figure 2,
StratUm extends the capabilities of the GJMF and
contributes (additional) abstractions for:
3.3.1 Efficient Service Communication
In addition to web service interfaces, StratUm pro-
vides access to the framework through a custom pro-
tocol called the Resource Access and Serialization
Protocol (RASP). RASP is a message-oriented wire-
transport hybrid protocol designed for efficient pars-
ing and serialization of message data. Messages are
represented in tree format, where tree nodes contain
hash maps that map text-resolved tags to binary data.
To support efficient transmission of large messages,
RASP supports both enveloped transmissions of large
binary payloads and chunked data transfer modes.
3.3.2 Data Management
GJMF employs a data management model where
GJMF coordinates third party data transfers, but does
not actively participate in data management. StratUm
extends the GJMF data management capabilities by
providing convenient interfaces for data monitoring
and control, an efficient protocol for data transmis-
sion, and caching and storage of staged data files.
3
In Geology, a stratum refers to a layer of sedimentary
rock. StratUm is designed to constitute an abstractive layer
for computation management. The Um emphasis of the
name refers to the place of origin, Ume
˚
a University.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
292
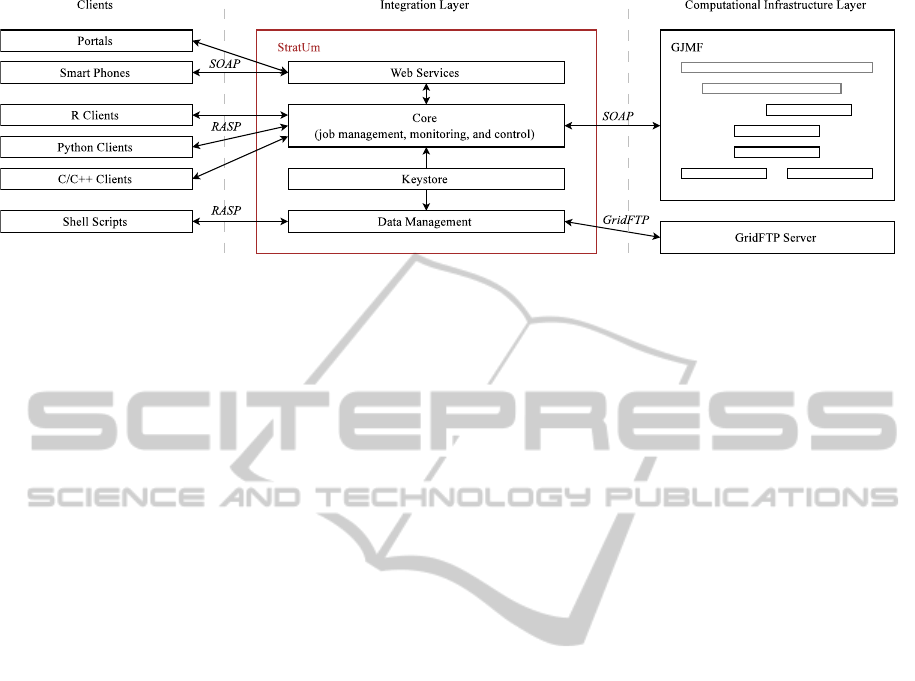
Figure 2: The StratUm architecture. Framework functionality exposed as services accessible through a custom protocol and
(optional) web services. Native client APIs abstract service communication, credentials, and data management complexity.
3.3.3 Security Models
GJMF utilizes a security model based on the GT4 im-
plementation of GSI where x509 certificate and del-
egated credential proxies are used for authentication
and authorization of end-users. StratUm provides
an extended security model that allows establishment
of secure communication channels using either cer-
tificates or username-password authentication (using
challenge-response message exchanges). In StratUm,
username-password tokens are associated to creden-
tials dynamically, which allows end-users to install,
remove, and update certificates and key pairs dynami-
cally during execution of computational tasks. In con-
junction with the GT4 credentials delegation mecha-
nism, StratUm supports creation and caching of dele-
gated certificates and key pairs.
3.3.4 Notification Models
Notification propagation via WSRF (as done in
GJMF) provides standardization of message formats
and programming models. WSRF is based on SOAP
web services and may impact integration complex-
ity (e.g., require clients to host SOAP containers) as
well as incur communication overhead in distributed
systems. For more efficient communication mod-
els and reduced integration complexity, StratUm en-
capsulates the GJMF notification model and provides
an out-of-band mechanism for asynchronous notifica-
tions based on propagation of status updates that are
transparently enveloped in RASP messages.
3.3.5 Native Client APIs
To facilitate client integration in end-user environ-
ments, StratUm provides a set of multi-language
client APIs for communication with StratUm ser-
vices. These client APIs define high-level interfaces
for job management as well as low-level mechanisms
for, e.g., message construction, that facilitate func-
tionality extension and framework customization. In
addition to client APIs in Java, C/C++, and Python,
StratUm also provides a set of command-line tools
that illustrate use of the StratUm client APIs and con-
stitute useful tools for distributed job management.
4 DISCUSSION
From the application standpoint, the presented system
greatly assists in large-scale scientific inquiry. De-
pending on the model under study and parameters
such as the time-horizon of the simulation and the
density of output samples, the nature of the individ-
ual tasks will vary from highly compute-intensive to
highly data-transfer intensive. As the computational
power is scaled up, more advanced applications such
as parameter estimation or optimization using, e.g.,
genetic algorithms will become computationally fea-
sible. In those cases, it is desirable to optimize re-
source access patterns to meet application-specific re-
quirements, e.g. minimizing the asynchrony of a task-
group or maximizing throughput for the SweGrid.
StratUm is well prepared for such extensions, and due
to the design and the well-defined API, application
specific workflow management can be added without
compromising the generality of the framework.
5 RELATED WORK
Design goals similar to those presented here are im-
plemented in the Virtual Infrastructure for simula-
tions with MCell, a software for microscale biochem-
ical simulation in grid environments (Casanova et al.,
2004). Mesoscale stochastic computations in a cloud
environment are demonstrated using the domain-
specific language Neptune (Bunch et al., 2011).
ABSTRACTIONS FOR SCALING eSCIENCE APPLICATIONS TO DISTRIBUTED COMPUTING ENVIRONMENTS
- A StratUm Integration Case Study in Molecular Systems Biology
293

A number of efforts similar to StratUm and GJMF
exist and include, e.g., Falkon (Raicu et al., 2007),
a lightweight task execution framework designed for
Many Task Computing (Raicu et al., 2008). Falkon
and StratUm are similar in use of custom protocols
and service interfaces, but while Falkon is designed
for high submission throughput StratUm is more fo-
cused on raising computation abstraction levels.
GridSAM (Lee et al., 2005) is a standards-based
grid job submission system that abstracts underlying
resource managers through a Web Service interface
built on staged event-driven architecture (SEDA). The
job submission pipeline of GridSAM is similar to the
integration of StratUm and GJMF, but the focus of
StratUm and GJMF lie more towards automation and
adaptability in system architecture design.
The Simple API for Grid Applications
(SAGA) (Kaiser et al., 2006) is an API stan-
dardization initiative that like StratUm and the GJMF
aims to provide a unified interface to grid integration.
The philosophy of the SAGA API differs from
StratUm in that StratUm focuses on automation
and minimization of integration complexity. Per-
formance evaluations comparing StratUm to SAGA
implementations are subject for future work.
For reasons of brevity, the list is far from com-
plete. Compared to most approaches, the aim of this
work is directed more towards design patterns for ab-
straction of complexity in computation management,
and exploration of approaches suitable for the specific
use cases of eScience applications. For more exhaus-
tive treatment of grid job management mechanisms
readers are referred to (
¨
Ostberg and Elmroth, 2010).
6 CONCLUSIONS
In this paper we explore an approach to integration
of eScience applications with distributed computing
environments based on separation of computation de-
sign from computation management. Emphasis of the
work is placed on architectures that raise computa-
tion management abstraction levels and facilitate ver-
satility in computation enactment. As part of the re-
sults we present StratUm, an integration architecture
designed to abstract complexity and raise abstraction
levels in distributed computation management.
ACKNOWLEDGEMENTS
The authors thank Mikael
¨
Ohman, Sebastian Gr
¨
ohn,
and Anders H
¨
aggstr
¨
om for work related to the
project. This work is done in collaboration with
the High Performance Computing Center North
(HPC2N) and is funded by the Swedish National In-
frastructure for Computing (SNIC), the Swedish Gov-
ernment’s strategic research project eSSENCE, the
Swedish Royal Academy of Sciences, and U.S NSF
Grant DMS-1001012, U.S. NIH Grant R01EB7511,
U.S. DOE Award DE-FG02-04ER25621, U.S. NSF
IGERT DGE-02-21715, Institute for Collaborative
Biotechnologies Grant DAAD19-03-D-0004 from the
U.S. Army Research Office.
REFERENCES
Bunch, C., Chohan, N., Krintz, C., and Shams, K. (2011).
Neptune: a domain specific language for deploy-
ing hpc software on cloud platforms. In Proceed-
ings of the 2nd international workshop on Scientific
cloud computing, ScienceCloud ’11, pages 59–68,
New York, NY, USA. ACM.
Casanova, H., Berman, F., Bartol, T., Gokcay, E., Se-
jnowski, T., Birnbaum, A., Dongarra, J., Miller, M.,
Ellisman, M., Faerman, M., Obertelli, G., Wolski, R.,
Pomerantz, S., and Stiles, J. (2004). The virtual in-
strument: Support for grid-enabled mcell simulations.
International Journal of High Performance Comput-
ing Applications, 18(1):3–17.
Drawert, B., Engblom, S., and Hellander, A. (2011). UR-
DME 1.1: User’s manual. Technical Report 2011-003,
Department of Information Technology, Division of
Scientific Computing, Uppsala University.
Foster, I. (2005). Globus toolkit version 4: Software for
service-oriented systems. In Jin, H., Reed, D., and
Jiang, W., editors, IFIP International Conference on
Network and Parallel Computing, LNCS 3779, pages
2–13. Springer-Verlag.
Kaiser, H., Merzky, A., Hirmer, S., Allen, G., and Seidel,
E. (2006). The saga c++ reference implementation:
a milestone toward new high-level grid applications.
In Proceedings of the 2006 ACM/IEEE conference on
Supercomputing, SC ’06, New York, NY, USA. ACM.
Lee, W., McGough, A. S., and Darlington, J. (2005). Per-
formance evaluation of the GridSAM job submission
and monitoring system. In UK e-Science All Hands
Meeting, pages 915–922.
¨
Ostberg, P.-O. and Elmroth, E. (submitted, 2010).
GJMF - A Composable Service-Oriented Grid Job
Management Framework. Preprint available at
http://www.cs.umu.se/ds.
Raicu, I., Foster, I., and Zhao, Y. (2008). Many-task com-
puting for grids and supercomputers. In Workshop on
Many-Task Computing on Grids and Supercomputers
(MTAGS) 2008., pages 1–11.
Raicu, I., Zhao, Y., Dumitrescu, C., Foster, I., and Wilde,
M. (2007). Falkon: a Fast and Light-weight tasK ex-
ecutiON framework. In Proceedings of IEEE/ACM
Supercomputing 07.
BIOINFORMATICS 2012 - International Conference on Bioinformatics Models, Methods and Algorithms
294
