
1-D MATHEMATICAL MORPHOLOGY FOR WATER
REMOVAL IN
1
H MR SPECTROSCOPY TOOL
Juan José Fuertes
1
, Valery Naranjo
1
, Jesús Angulo
2
and Mariano Alcañiz
1,3
1
Instituto Interuniversitario de Investigación en Bioingeniería y Tecnología Orientada al Ser Humano (I3BH),
Universitat Politècnica de València, I3BH/LabHuman, Camino de Vera s/n, 46022 Valencia, Spain
2
Centre de Morphologie Mathématique, Mathématiques et Systèmes, MINES Paristech, Paris, France
3
Ciber, Fisiopatología de Obesidad y Nutrición, CB06/03 Instituto de Salud Carlos III, Madrid, Spain
Keywords: Magnetic Resonance Spectroscopy, Signal Processing, Brain Cancer Detection, Multivoxel, Magnetic
Resonance Imaging.
Abstract: This work shows the basics and performance of a new morphological signal method for 1-D water signal
removal included in a simple and interactive multivoxel spectroscopy tool to help surgeons detect brain
cancer. It consists of mathematical morphology usually applied in 2D images to filter 1D spectroscopic
signals. 1D water signal reconstruction from the original data is performed in frequency domain through the
use of an elementary operation: geodesic dilation. Then, the water signal is subtracted from the original
signals due to the large amount of water which exists in the brain compared to the rest of molecules, making
possible quantitation procces. The goal of this paper is to present this new morphological method commonly
used in 2D domain for 1D water removal, spreading its use to several processing methods as quantitation.
1 INTRODUCTION
Many spectroscopic algorithms have been developed
to solve the problem of quantifying signals in
1
H
MRS data in the last 20 years, being them included
in the most popular software tools, such as the
jMRUI software package (Stefan, Di Cesare,
Andrasescu, Popa, Lazariev, Vescovo, Strbak,
Williams, Starcuk, Cabanas, Van Ormondt and
Graveron-Demilly, 2009) and the AQSES software
(Simonetti, Poullet, Sima, De Neuter, Vanhamme,
Lemmerling and Van Huffel, 2006). One of the most
important algorithms in
1
H MRS is the water
suppression from original signals, in order to
eliminate the large amount of water which exists in
the brain compared to the rest of molecules. The so-
called blackbox methods based on singular value
decomposition (SVD) such as HSVD (De Beer and
Van Ormondt, 1992), HLSVD (Pijnappel, Van den
Boogaart, De Beer and Van Ormondt, 1992) and
HTLS (Van Huffel, Chen, Decanniere and Van
Hecke, 1994), have been successful in
reconstructing the signal as a sum of Lorentzians,
but the influence that the users have when the
estimated parameters are chosen could reduce the
quality of the signal fitting. Another important
method is the maximum-phase Finite Impulse
Response (MP-FIR) which has been shown to be
one of the most accurate and efficient technique for
quantifying MRS spectra (Sundin, Vanhamme, Van
Hecke, Dologlou, and Van Huffel, 1999). In
addition, MP-FIR allows the inclusion of prior
knowledge that may be taken into account during
quantitation; however the drawback of these filters is
that they are linear phase filters which generate
distortion due to the fact that the signals are
composed of exponentially damped sinusoids and
not pure sinusoids, and this distortion cannot be
totally neglected. Other techniques as Wavelets
(Günther, Ludwig, and Rüterjans, 2002) or Gabor
transforms (Coron, Vanhamme, Antoine, Hecke, and
Van Huffel, 2001) have also used for water removal
but with lower accuracy than the other ones. These
methods are compared in a filtering review to solve
suppression in MRS (Coron et al., 2001): the MP-
FIR method was the most accurate and efficient
technique. The water suppression method presented
in this paper belongs to a general process where the
main steps included in most of spectroscopy
software tools are illustrated in Figure 1:
381
Fuertes J., Naranjo V., Angulo J. and Alcañiz M..
1-D MATHEMATICAL MORPHOLOGY FOR WATER REMOVAL IN 1H MR SPECTROSCOPY TOOL.
DOI: 10.5220/0003770403810384
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2012), pages 381-384
ISBN: 978-989-8425-89-8
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

Figure 1: Block diagram for
1
H MR spectroscopy
analysis.
In this work we are focusing on the dominant
signals removal, specifically in the water
suppression algorithm. This method is included in a
software tool (See Figure 2) which also incorporates
the main algorithms to process data, performs the
registration between spectroscopic data and MR
images, and generates the metabolite maps. In short,
the aim of this paper is to provide researchers with a
new morphological method for water suppression,
introducing it for a future use in quantitation
metabolite signals.
This paper is set up as follows: section 2
reminds some basics of morphology and the main
characteristics of mathematical algorithms: dilation,
erosion, opening and closing. In section 3, the water
removal algorithm based on mathematical operations
is shown. Section 4 explains the experiments and
finally, a brief conclusion is given in section 5
together with the future work.
Figure 2: Main window of the
1
H MR Spectroscopy tool.
2 MATHEMATICAL
MORPHOLOGY
Mathematical morphology is a non-linear
image/signal processing methodology based on
minimum and maximum operations (Serra, 1982), in
order to extract relevant components of an
image/signal respectively. In the last years, this
technique has been commonly used in 2D images,
but its use for 1D signal processing is also possible.
Let f be a signal which is defined as:
: → (1)
where
∈ is the time sample. In the case of
discrete valued signals, =
,
1,…
is an ordered set of amplitude-levels.
Furthermore, let
be a sub-set of called
structuring element (shape probe) centred at
point
. The size of the structuring element (SE) is
usually chosen according to some a priori
knowledge about the geometry of the relevant and
irrelevant signal components. The two basic
morphological operators are:
• Dilation:
=
∈
• Erosion:
=
∈
Those elementary operations can be combined to
obtain a new set of operators or basic filters given
by:
• Opening:
=
• Closing:
=
There are other complex filters derived from the
basic operator based on geodesic transformations of
a signal f (marker) and a second signal g (reference):
• Geodesic reconstruction:
=
with
=
⋀
• Opening by reconstruction:
=
with
=
.
A combination of these operators can be used if
we want to reconstruct the water signal and subtract
it from the original signals as it has been introduced
in section 1. In next section the basic morphological
operation will be illustrated.
3 WATER SIGNAL SUPPRESSION
In order to perform water removal, many algorithms
have been proposed since 1990 in the time and
Patient’s Brain
MR Imaging
Ac
q
uisition
Signal
Ac
q
uisitions
Quantitation
Phase Correction
Si
g
nal Enhancement
Water Removal Parameter Estimation
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
382
frequency domain (Vanhamme, Sundin, Hecke and
Huffel, (2001); Poullet, Sima and Huffel, (2008)),
but nowadays only some of them are really used in
multivoxel spectroscopy imaging. Ideally, the FID
signal obtained with the MRS machine is noiseless
and results from the addition of K exponentially
damped sinusoids which are characterized by
frequencies f
k
, amplitudes A
k
in arbitrary units (a.u.),
phases φ
k
, damping factors α
k
, length of the FID N, i
the square root of -1 and Δt the sampling interval, as:
X
=
∑
A
e
e
n= 0,...,N-1. (2)
In order to remove the water signal, the
exponentially damped sinusoids whose frequencies
appear in the water region are estimated and
subtracted from the original FID. For example,
Hankel Lanczos Singular Value Decomposition
(HLSVD-PRO) algorithm (Laudadio, Mastronardi,
Vanhamme, Van Hecke and Van Huffel, 2002),
estimates the whole set of model parameters making
full use of mathematical model functions,
reconstructing the water signal and subtracting it
from the original signals.
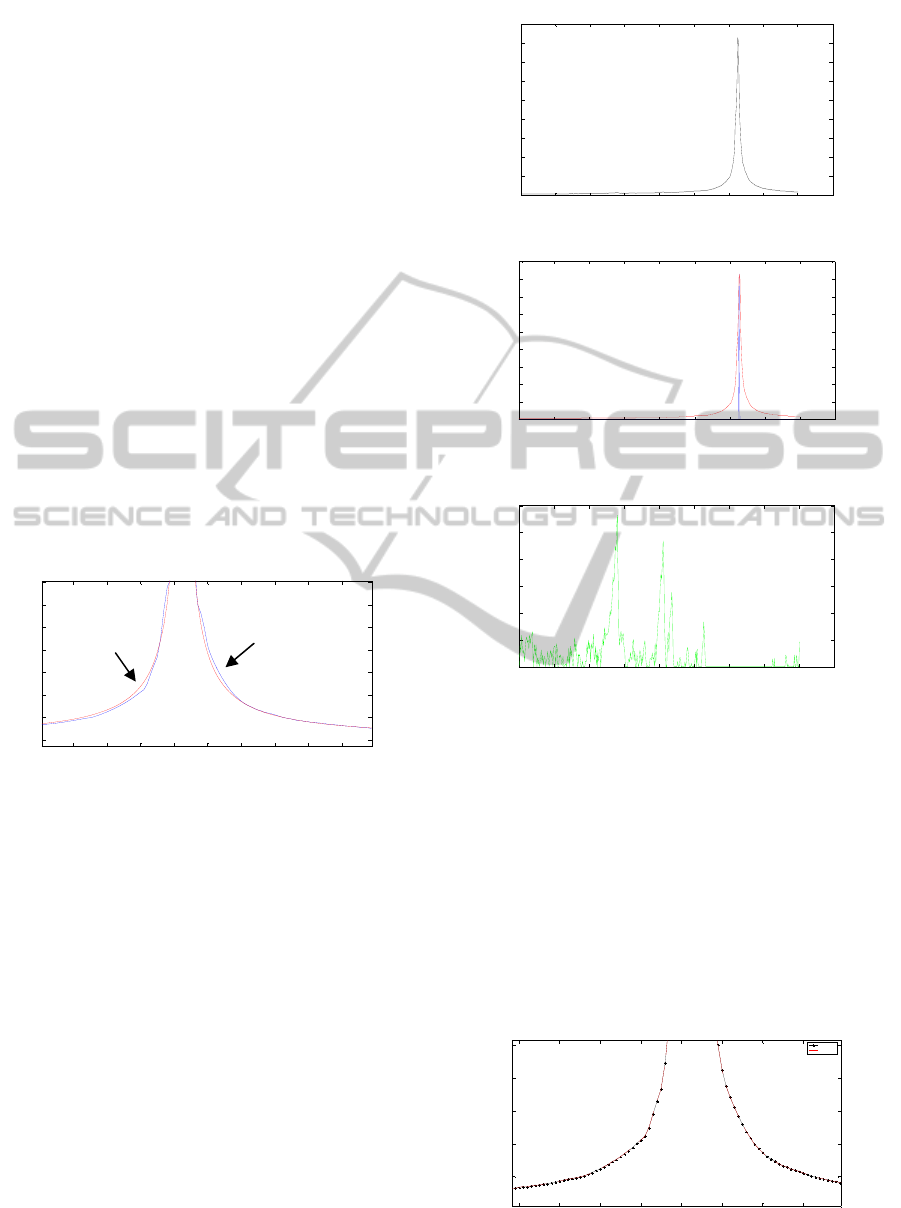
Figure 3: Imperfect reconstruction of water contribution to
subtract it from the original signal. In blue and in red the
original and the reconstructed signal respectively.
The main problem of this method lies in the
accurate reconstruction of the water contribution to
subtract it from the original signal (Figure 3);
sometimes the water peak is not well reconstructed,
other times the base of a signal has imperfections
and it is so difficult to fit the parameters. For this
reason, the method proposed in this work uses the
geodesic reconstruction by means of a delta centred
on the maximum value of the original signal as
marker, and the original signal as reference signal,
allowing us to perform the water reconstruction
accurately in frequency domain without affecting
other signals. In figures 4, 5, and 6, the full process
of water suppression is illustrated.
Figure 4: The original spectroscopic signal.
Figure 5: In blue, the delta used as marker. In red, the
reconstructed signal after using geodesic reconstruction.
Figure 6: Residue after performing the process.
4 EXPERIMENTS AND RESULTS
As it was told in section 2, the introduction of
mathematical morphology for 1D signal processing
could help us when we want to suppress water signal
or calculate metabolite quantitation. In Figure 7 the
original spectroscopic signal in black and the water
reconstruction in red using the geodesic
reconstruction are observed together. Then, the red
signal is subtracted from the original one in time
domain, eliminating the large amount of the water.
Figure 7:
1
H MR original spectroscopy (black) and water
reconstruction (red) signals.
480 490 500 510 520 530 540 550 560
0
0.5
1
1.5
2
2.5
3
3.5
x 10
4
0 50 100 150 200 250 300 350 400 450
0
1
2
3
4
5
6
7
8
9
x 10
4
O riginal
0 50 100 150 200 250 300 350 400 450
0
1
2
3
4
5
6
7
8
9
x 10
4
M arcador (azul) y Reconstruida (rojo)
0 50 100 150 200 250 300 350 400 450
0
100
200
300
400
500
600
Solo el residuo original-reconstruida (verde)
470 480 490 500 510 520 53 0 540
0.5
1
1.5
2
2.5
x 10
4
original
reconstructed
f (points)
Amplitude (a.u.)
Amplitude (a.u.)
Amplitude (a.u.)
Amplitude (a.u.)
f (points)
f (points)
f (points)
f (points)
Amplitude (a.u.)
1-D MATHEMATICAL MORPHOLOGY FOR WATER REMOVAL IN 1H MR SPECTROSCOPY TOOL
383
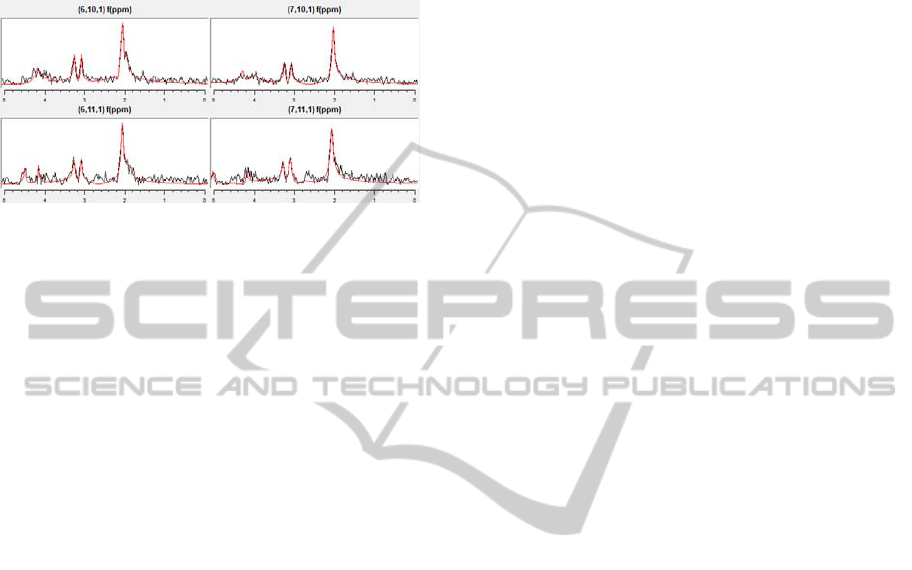
Other processing algorithms as phase correction,
apodize funtions or SNR improvement can be
applied. Figure 8 shows the result after signal
filtering, SNR improvement and HLSVD
quantitation for several spectroscopic signals.
Figure 8: Signal processing for metabolite quantitation.
5 CONCLUSIONS AND FUTURE
WORK
In this paper, we have introduced the use of 1D
mathematical morphology in a software tool to help
surgeons detect brain cancer through the use of the
magnetic resonance spectroscopy. This method is
appropriate for water removal when the metabolite
signals are not overlapped with the water
contribution; otherwise, methods as HLSVD are
recommended in order to avoid a bad suppression.
However, a depth study about the use of
morphological methods in overlapped signals must
be done since an appropriate family of filters can
obtain the desired signal. Coming soon experiments
are focused on the comparison of the proposed
algorithm with other filtering methods to verify its
efficiency and in the study of new quantification
methods based on non-linear filters (1D
mathematical morphology) for its use in MRS.
ACKNOWLEDGEMENTS
This work has been supported by Centro para el
Desarrollo Tecnológico Industrial (CDTI) under the
project ONCOTIC (IDI-20101153), and partially by
projects Consolider-C (SEJ2006-14301/PSIC),
“CIBER of Physiopathology of Obesity and
Nutrition, an initiative of ISCIII” and Excellence
Research Program PROMETEO (Generalitat
Valenciana. Conselleria de Educación, 2008-157).
We would like to express our deep gratitude to the
Hospital Clínica Benidorm for its participation in
this project. The work of Juan José Fuertes has been
supported by a FPI grant from “Programa de Ayudas
de Investigación y Desarrollo (PAID)” of UPV.
REFERENCES
Coron, A., Vanhamme, L., Antoine, J. P., Hecke, P. V.
and Van Huffel, S. (2001). The filtering approach to
solvent peak suppression. Journal of Magnetic
Resonance. 152(1):26-40.
De Beer, R. and Van Ormondt, D. (1992). Analysis of
NMR data using time domain fitting procedures. NMR
Basic Principles and Progress. 26:202-48.
Günther, U. L., Ludwig, C. H., and Rüterjans, H. (2002).
WAVEWAT-improved solvent suppression in NMR
spectra employing wavelet transforms. Journal of
Magnetic Resonance. 156(1):19-25.
Laudadio, T., Mastronardi, N., Vanhamme, L., Van
Hecke, P. and Van Huffel, S. (2002). Improved
Lanczos algorithms for blackbox MRS data
quantitation. Journal of Magnetic Resonance.
157:292-297.
Pijnappel, W. W. F., Van den Boogaart, A., De Beer, R.,
and Van Ormondt, D. (1992). SVD-based
quantiffication of magnetic resonance signals. Journal
of Magnetic Resonance. 97(1):122-134.
Poullet, J. B, Sima, D. M., and Van Huffel, S. (2008).
MRS signal quantitaion: a review of time- and
frewuency-domain methods. Journal of Magnetic
Resonance. 195(2):134-144.
Serra, J. (1982). Image Analysis and Mathematical
Morphology, volume I. Ac. Press, London.
Simonetti, A. W., Poullet, J-B., Sima, D. M., De Neuter,
B., Vanhamme, L., Lemmerling, P., and Van Huffel S.
(2006). An open source short echo time MR
quantitation software solution: AQSES.
Stefan, D., Di Cesare, F., Andrasescu, A., Popa, E.,
Lazariev, A., Vescovo, E., Strbak, O., Williams, S.,
Starcuk, Z., Cabanas, M., Van Ormondt, D., and
Graveron-Demilly, D. (2009). Quantitation of
magnetic resonance spectroscopy signals: the jMRUI
software package. Measurement Science and
Technology, volume 20.
Sundin, T., Vanhamme, L., Van Hecke, P., Dologlou, I.,
and Van Huffel, S. (1999). Accurate quantification of
1H spectra: From infinite impulse response filter
design for solvent suppression to parameter
estimation. Journal of Magnetic Resonance.
139(2):189-204.
Van Huffel, S., Chen, H., Decanniere, C., and Van Hecke,
P. (1994) Algorithm for time-domain NMR data
fitting based on total least squares. Journal of
Magnetic Resonance. 110:228-237.
Vanhamme, L., Sundin, T., Hecke, P. V., and Huffel, S.
V. (2001). MR Spectroscopic Quantitation: a Review
of Time-Domain Methods. NMR in Biomedicine.
14:233-246.
NAA
Cr
Cho
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
384
