
AN OPENEHR REPOSITORY BASED ON A NATIVE XML
DATABASE
Linda Velte
1
, Tiago Pedrosa
2
, Carlos Costa
1
and José Luís Oliveira
1
1
University of Aveiro, DETI/EETA, Aveiro, Portugal
2
Polytechnic Institute of Bragança, 5301-854, Bragança, Portugal
Keywords: EHR, openEHR, XML repository.
Abstract: openEHR is an open standard specification that describes the management, storage, retrieval and exchange
of data in Electronic Health Record (EHR). Despite its growing importance in the field, the lack of open
source solutions is hindering a larger visibility. In this paper we present an openEHR-based repository
supported by a native XML database, which allows to store and query openEHR records through the DB
service layer and a set of REST web services. The obtained results highlight the efficiency of this API and
show that it can be used as a persistence component in any openEHR solution.
1 INTRODUCTION
An EHR allows an integrated access to the patients
information as it gives the possibility to aggregate
all medical data of the patient, and consequently, to
have a patient centric storage approach. A patient’s
EHR must be accessible and the information
understandable, no matter what hospital/medical
institution he visits. To accomplish that, there must
be an agreement on the language that is ”spoken”.
This is done via standards that define not only how
the information is structured and represented but
also how it can be retrieved and shared between
systems (Sunyaev, 2008). Existing standards can be
divided into two major groups: 1) to define the
content format, like, for instance, openEHR, MML
(Medical Markup Language) and HL7 CDA (Health
Level 7 Clinical Document Architecture) and 2) to
define how the communication should be done like,
for instance, WADO (Web Access to DICOM
Persistent Objects), RID (Retrieve Information for
Display) and XDS (Cross-Enterprise Document
Sharing). Some standards can be included in both of
these groups (DICOM Structured Reports and
EHRcom) (MITRE, 2006). Within these standards, a
great expectation is being put on openEHR, which
usage has been increasing over the years (Eicheberg,
2005). Most standards focus on a specific area, such
as DICOM focuses on digital imaging or HL7 on
patients’ administration. On other hand, openEHR
has the goal to provide semantic interoperability
between all medical specialties, reducing the needed
of standards to a single one, in opposition to the
several standards that are used nowadays.
An important aspect in openEHR solutions is the
storage technology. This kind of repository needs a
very generic database model, which is not supported
by current implementations. Opereffa project
(Opereffa, 2011) is an openEHR repository that uses
a relational database, with just one table to store
path/value pairs for each attribute. A strong
drawback of this approach is that for patient centric
queries the information has to be reassembled via
complex joins operations.
In this paper we propose a repository supported
by a XML database, where each patient has all his
information in one record, simplifying storage and
query procedures.
2 OPENEHR
OpenEHR is an open standard specification in health
informatics that describes the management, storage,
retrieval and exchange of data in EHR’s (openEHR,
2011). In this standard, all data relative to a person is
stored in a ”one lifetime” vendor independent and
centred EHR (Kalra, 2005).
The goal of openEHR standard is to achieve
semantic interoperability regarding the whole health
386
Velte L., Pedrosa T., Costa C. and Oliveira J..
AN OPENEHR REPOSITORY BASED ON A NATIVE XML DATABASE.
DOI: 10.5220/0003784003860389
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2012), pages 386-389
ISBN: 978-989-8425-88-1
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

area. For this, a generic, dynamic health information
model is needed, capable of storing new kinds of
data without having to change the data model. The
drawback of this kind of model is that there is no
control about what information is stored, leading to
low data quality and is not very different from an
unstructured model. This issue was solved by an
innovative modeling approach that creates a second
level used to constrain the information (Bird, 2003).
This decoupling methodology consists basically in
the separation of information and knowledge. The
result is a generic health record model that enables
the storage of a wide variety of health information.
The openEHR architecture can be divided into
two major parts: Reference Model the Archetype
Model. The former allows interoperability because
data is exchanged between systems only in terms of
standard open reference model instances; the latter
allows semantic interoperability. Additionally, there
is a Service Model includes definitions of basic
services in the health information environment,
which are EHR centred.
The structure of the information is constrained
by the archetype model, which defines exactly
which data types, structures and values are valid,
making the information both flexible and structured.
An archetype represents a clinical concept and it is
used to constrain instances of the openEHR
information model by defining a valid structure, data
types and values. An electronic health record that
has been archetyped will have the same meaning no
matter where it appears, allowing it to be shared by
multiple health systems (Beale, 2002). Archetypes
are defined using the Archetype Definition
Language (ADL) (Beale and Heard 2005). To query
openEHR information it was defined the Archetype
Query Language (AQL) that allows to access the
nodes in a similar way as a XPath query on an XML
document (Beale, 2005).
There is already available a fully implemented
version of the reference and archetype models – the
openEHR Java Reference Implementation (Chen,
2007) – and many organizations are using or
contributing to the openEHR standard. Two major
references are Opereffa (Opereffa, 2011) and
Kanolab (Kanolab, 2011).
3 A OPENEHR REPOSITORY
The purpose of our repository is to store electronic
health records according to the openEHR standard,
offering an API to perform database management
functions (like insert, edit or delete a record) and to
query the inserted records. To accomplish this task,
a few aspects have to be considered. A very
important one is the fact that the repository must
store openEHR records using a dynamic database
model. Another fact to have in consideration is that
the repository is only a part of a healthcare system
and, consequently, has to be inserted into this system
architecture. To make this possible the
implementation should have a well-defined set of
services that are reusable by any application. Those
services should provide the functionalities normally
supported by a healthcare information system,
namely operations related with records search and
management.
The system architecture was designed to divide
functionalities into three different layers, following
the Service Oriented Architecture (SOA) model
(Papazoglou, 2003). This approach has the
advantage of providing reusable services, being
possible to integrate implemented functionalities in
any application. The result is shown in Figure 1.
Figure 1: System Architecture.
3.1 Core Layer
The Core layer is responsible for managing the
information to be stored. The storage technology
used was a native XML database, a relatively new
technology and, consequently an interesting case of
study, namely to compare performance with
AN OPENEHR REPOSITORY BASED ON A NATIVE XML DATABASE
387

relational approaches or with non-native XML
databases (i.e. DBs that use a relation model with
XML fields to store the data).
XML databases perform better when used in
document centric applications, what makes it perfect
to support a patient centric EHR repository. This
kind of storage approach makes possible to have a
XML file per patient containing all his medical data.
For our implementation we have used BaseX an
open-source and platform-independent XML DB,
written in Java, with an Xpath/XQuery processor,
and with support for W3C Update and Full Text
extensions. The W3C Update is a great advantage
regarding performance issues, allowing us to access
to a specific position of the file to read or modify its
value. Normally, other solutions obligate to read the
entire file, change it and rewrite it. Moreover, it
offers a RESTful API for accessing distributed XML
resources, which provides a simple and fast access to
databases though HTTP (using HTTP methods GET,
PUT and DELETE).
The patient repository consists in one unique
XML node that contains the EHR records, following
the openEHR structure (Figure 2).
Figure 2: High Level Structure of the openEHR (openEHR
2011).
Each object is identified by an EHR ID and
contains structured information that is versioned,
plus a list of Contribution objects that act as audits
for changes. The EHR Access and EHR Status
objects contain control information, while the
Directory (optional) can be used to hierarchically
organize Compositions, objects that contain the
patient medical information. Every EHR has also a
list of Contributions that are used to save every
change made to the record.
3.2 Service Layer
The set of implemented services allow the remote
and transparent management of electronic health
records storage and the query of information
contained in these records. All the implemented
services were implemented using SOAP and their
functionalities can be divided into four groups
according to their application area: DBManager,
ServerManager, EHRManager and QueryService.
The DBManager module is responsible for the
administrative operations on the repository, such as
creating a new database or obtaining information
about an already existing database. The
EHRManager is responsible for all the Web Services
related to record manipulation. This set of services
cover the requisites regarding the information
management of electronic health records, such as
insertion, edition, or deletion of EHRs, compositions
and contributions. The QueryService allows
querying the data available in the repository. The
two available services to retrieve information are
executeQuery (free XPath/XQuery queries) and
textQuery (full text search). The ServerManager file
contains the services responsible for the server
management, such as starting or stopping the BaseX
server.
3.3 Application Layer
To validate the functionalities of the developed
openEHR repository we have also developed a web
application with a particular set of client services,
namely: a) insert/edit/remove EHRs; b) view EHR
content; c) get Composition list associated to an
EHR; d) insert/edit/remove Compositions; e) query
database (XPath and free text).
The application was developed in ZK, an open-
source web solution written in Java that includes an
Ajax-based event-driven engine and a rich set of
components. The Web application works as an
administration platform for any repository.
4 EVALUATION
To evaluate the performance of developed openEHR
Repository systematic tests were made for series of
1000, 10000 and 30000 patient records. Several
operations were analysed, included: a) Add a new
EHR to the database; Search for a patient record;
Search for an attribute; Add a composition to an
existing record.
In our openEHR repository, the query processing
is notably faster than the insertion process.
Analysing the context where the repository will be
used (health information systems), it is more
important to have faster queries than to have faster
insertions (add a new patient or add new data to the
HEALTHINF 2012 - International Conference on Health Informatics
388
patient’s record). After the tests we have concluded
that while the query time for a record is independent
of the volume of the database (approximately
0.65ms), the insertion time increases with the
numbers of records already inserted (with 30000
records inserted, it took 423ms to insert an
additional one). The major conclusion is that this
repository is best suitable for systems that require a
fast query response to patient centric queries.
Using the index system mechanism offered by
BaseX it was possible to obtain the same response
time to query a patient’s medical record,
independently of the number of records inserted. It
also made no difference which position the record
occupies in the repository file.
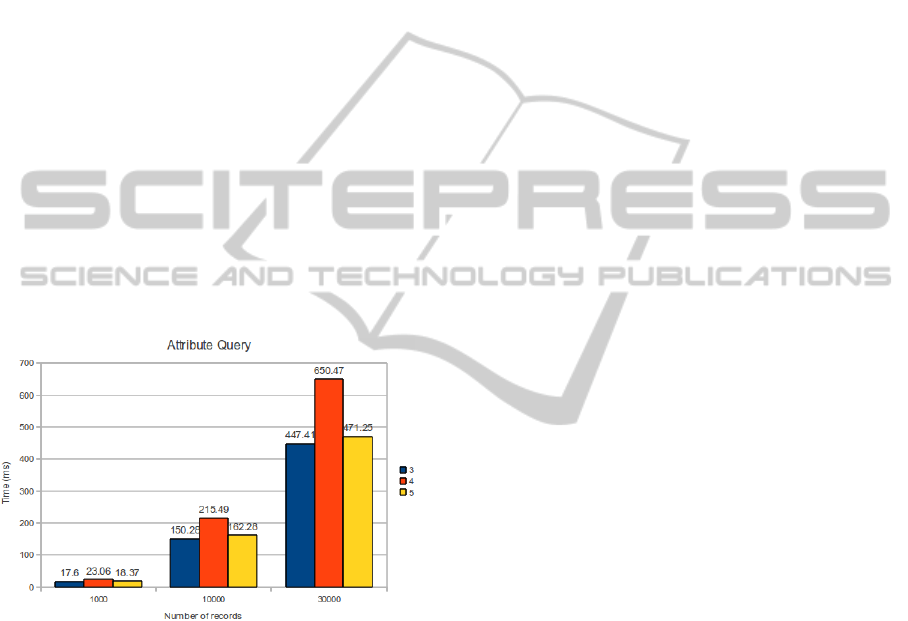
Information centric queries, i.e. search for
special attributes along a set of patients, are much
slower and depend on the number of records inserted
(Figure 3). In our approach, queries like, for
instance, “how many patients have high blood
pressure?” obligates to analyse the whole
information inserted. To cope with this we need to
deploy a more extensive indexing solution based on
Lucene, for instance.
Figure 3: Attribute Search.
5 CONCLUSIONS
In this paper we have presented a generic and
dynamic database, able to store electronic health
records according to the openEHR standard.
Additionally, the developed SOAP API offers a
unified access to the repository, making it possible
to integrate it with any kind of application. The goal
is to promote the development of health information
systems based on the openEHR standard, allowing
an integration of all clinical areas (e.g. radiology,
administration, pharmacy) and making the process
more unified. Besides the repository and its clinical
usage, we have also developed a set of web services
and an administration web application that allow
patients to enter their own medical information into
secure health records. This support to PHR (Personal
Health Records) systems is an important asset,
considering the document centric approach of our
solution – every patient has all his information
stored in just one record.
The repository created and the services provided
enable the creation of client applications in an easy
way. The use of web services and the format of the
record stored on XML allow the use of generic
technology and programing languages.
REFERENCES
Beale T., 2002. Archetypes: Constraint-based domain
models for future-proof information systems.
OOPSLA 2002 workshop on behavioural semantics.
Beale, T., 2005. Archetype query language (aql),
openEHR specification, openEHR foundation.
Beale, T., Heard, S., 2005. Archetype definition language
(adl), openEHR specification, openEHR foundation.
Bird, L., Goodchild, A., Tun, Z., 2003. Experiences with a
two-level modelling approach to electronic health
records, Journal of Research and Practice in
Information Technology, 35(2):121.
Chen, R., Klein, G., 2007. The openehr java reference
implementation project. In Medinfo 2007, Proceedings
of the 12th World Congress on Health (Medical)
Informatics, Building Sustainable Health Systems,
page 58. IOS Press.
Eichelberg, T. Aden, Riesmeier, J., Dogac, A., Laleci
G.B., 2005. A survey and analysis of electronic
healthcare record standards, ACM Computing Surveys
(CSUR), 37(4):277–315.
Kalra, D., Beale, T., Heard, S, 2005. The openEHR
foundation, Studies in Health Technology and
Informatics, 115:153–173.
Kanolab, 2011. http://www.kanolab.info/index.php
Papazoglou, M. P., Georgakopoulos, D., 2003. Service-
oriented computing. Communications of the ACM, vol.
46 (10).
MITRE Coorporation, 2006. Electronic Health Records
Overview. National Institutes of Health National,
Center for Research Resources.
openEHR, 2011. http://www.openehr.org.
Opereffa, 2011, http://opereffa.chime.ucl.ac.uk/introducti
on.jsf
Sunyaev, A., Leimeister, J. M., Schweiger, A., Krcmar,
H., 2008. IT-Standards and Standardization
Approaches in Healthcare. Encyclopedia of
Healthcare Information Systems. 813-820.
AN OPENEHR REPOSITORY BASED ON A NATIVE XML DATABASE
389
