
LONG TERM BIOSIGNALS VISUALIZATION AND PROCESSING
Ricardo Gomes
1
, Neuza Nunes
2
, Joana Sousa
2
and Hugo Gamboa
1,2
1
Physics Department, FCT-UNL, Lisbon, Portugal
2
PLUX Wireless Biosignals S.A., Lisbon, Portugal
Keywords:
Biosignal, signal processing, Long term monitoring, Data structure.
Abstract:
Long term acquisitions of biosignals are an important source of information about the patients’ state and its
evolution, but involves managing very large datasets, which make signal visualization and processing a com-
plex task. To overcome these problems, we introduce a new data structure to manage long term biosignals.
A fast and non-specific multilevel biosignal visualization tool based on the concept of subsampling is pre-
sented, with focus on the representative signal parameters (mean, maximum, minimum and standard deviation
error). The visualization tool enables an overview of the entire signal and a more detailed visualization in
specific parts which we want to highlight. The ”Split and Merge” concept is exposed for long term biosig-
nals processing. A processing tool (ECG peak detection) was adapted for long term biosignals. Several long
term biosignals were used to test the developed algorithms. The visualization tool has proven to be faster
than the standard methods and the developed processing algorithm detected the peaks of long term ECG sig-
nals fast and efficiently. The non-specific character of the new data structure and visualization tool, and the
speed improvement in signal processing introduced by these algorithms makes them useful tools for long term
biosignals visualization and processing.
1 INTRODUCTION
The increasing development of medical systems and
applications for human welfare has been supported
by patients’ body signals monitoring. These biosig-
nals give the researcher or clinician a perspective over
the patient’s state since they carry information about
complex physiologic mechanisms. Biomedical signal
analysis has great importance for data interpretation
in medicine and biology.
In order to analyze the patient’s condition it is
very important to visualize the acquired signals and
extract relevant information from them. In clinical
cases such as neuromuscular diseases and sleep disor-
ders, a constant monitoring of the patient’s condition
is necessary (Pinto et al., 2010; Kayyali et al., 2008),
due to the possible occurrence of sudden alterations
in the patient’s state. However, long term acquisitions
generate large amounts of data, which exceed the ca-
pabilities for which standard analysis and processing
software were designed. In addition to the difficulty
of handling large amounts of data, displaying these
biosignals using standard visualization software is not
feasible due to our inability to correctly visualize the
entire signal. In a future perspective, the continuous
monitoring of biosignals will allow to know before-
hand when the patient needs assistance, assuring the
patients’ comfort as they are monitored in ambient as-
sisted living conditions (Sousa et al., 2010).
We present new tools for the visualization and pro-
cessing of very large biological datasets. The follow-
ing section presents the developed tools and the new
data structure for long term biosignals. In section 3
we present the methods of the developed work and
discuss the results and algorithm’s performance. Fi-
nally, we conclude the work in section 4.
2 PROPOSED DATA STRUCTURE
AND DEVELOPED TOOLS
As we are dealing with very long signals, a tool to
display large amounts of data is needed. Since we
used acquisition equipment that saves (raw) data in
text files, random access to a specific time window
of the recording wasn’t possible. To overcome this,
we created a new data structure that enables fast data
accessing, based on the HDF5 file format, a powerful
tool for managing different types of data (HDF group,
2007).
402
Gomes R., Nunes N., Sousa J. and Gamboa H..
LONG TERM BIOSIGNALS VISUALIZATION AND PROCESSING.
DOI: 10.5220/0003784704020405
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2012), pages 402-405
ISBN: 978-989-8425-89-8
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)
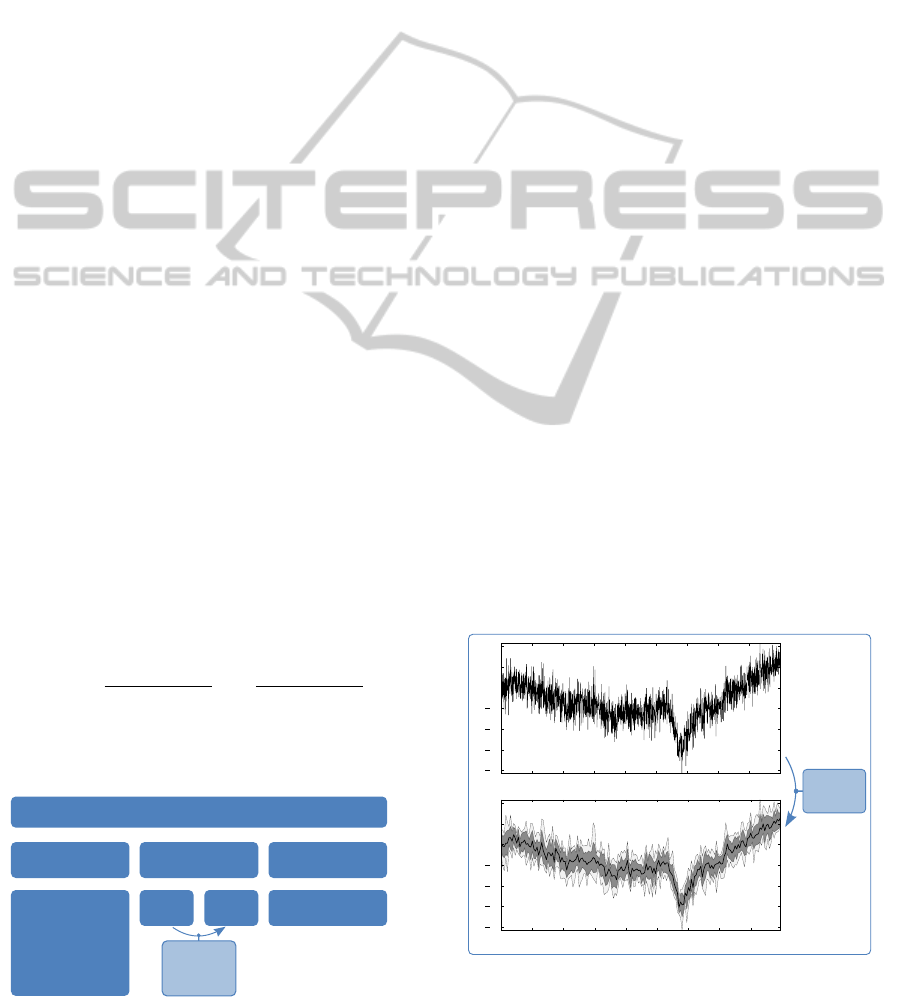
2.1 Long Term Biosignals Data
Structure
The data structure architecture (Figure 1) is based on
three sections: the acquisition data, the biosignals,
and the processed data. The biosignals section is com-
posed by the raw data and the different ”zoom levels”.
To obtain the different zoom levels, the four subsam-
pling parameters shown in Figure 1 are extracted from
the signal. Data mean identifies its central location,
being a representative measure of the signals’ shape.
Maximum and minimum parameters define the enve-
lope on which the signal is restrained, while the stan-
dard deviation error provides information about the
signal’s spreading.
The first (most detailed) level of visualization is
the raw data. The subsequent zoom levels provide
less detail than the preceding one, since they have a
smaller number of samples. However, they represent
the same time interval. The different zoom levels are
created by a subsampling process. Each subsampling
operation is carried out by splitting the input signal
in groups with a selected number of samples - the re-
sampling factor (r), and for each group the represen-
tative signals measures are calculated. The r factor
can be, for example 10, which means that the signal’s
measures will be computed from 10 to 10 samples.
Therefore, the data length will be divided by r, since
each group with r samples is represented by one new
sample. The first zoom level is obtained taking the
raw data as input. For higher zoom levels, the al-
gorithm takes as input the data from the last zoom
level to be created, taking advantage of the data reduc-
tion on each iteration, since it calculates the mean of
means, the maximum of maxima, minimum of min-
ima (calculating the mean is faster for 1000 values
than for 10000). The standard deviation error, (sd), is
obtained with equation1, where E[X] is the expected
value for the random variable X.
sd(X) =
q
E[X − E[X]
2
] =
q
E[X] −E[X]
2
(1)
The visual effect and data mining of the described
subsampling technique are shown in Figure 2.
biosignals
visualiza
zoom
levels
raw
data
processed data
.h5 file architecture
acquisition data
- subject's name
- date
- mac address
- sampled channels
- digital channels
- recording duration
- sampling frequency
- ECG peaks
(...)
subsampling
mean
maximum
minimum
standard error
Figure 1: Proposed data structure for biosignals.
2.2 Long Term Biosignals Visualization
Based on the presented data structure, a tool to vi-
sualize long term biosignals was implemented. This
tool provides an overview of entire long term signals
in the first instance and allows to zoom in and out
to specific time windows. This approach is compa-
rable to web mapping services, but applied to the vi-
sualization of electrophysiological signals. A client-
server model was implemented, giving the tool higher
portability, using Python as a way to manage data and
Javascript and HTML to create the visualization plat-
form. The initial display is done by drawing the entire
signal: the outermost (lowest) zoom level; the signal
being shown is updated when the navigation keys (for
zooming and panning) are pressed. Signal navigation
is facilitated by an overview window, that indicates
the selected region of the signal and enables the user
to select precise time windows in the signal to be visu-
alized in detail. There are two drawing stages which
allow a fast view of the signal’s shape:
• Preview. the signals’ informations to be drawn
are only the maximum and minimum (aiming for
a fast and representative overview);
• Detailed View. draws the signal’s mean, maxi-
mum, minimum, and the error shade (defined by
mean±standard deviation error).
These drawing steps enable a faster navigation,
since the user can ask for new time windows to be
displayed almost instantly. The detailed data is shown
only when the viewer stops in a specific time window,
providing the complete information about the signal.
When the raw data level is reached, no detailed infor-
mation is shown, since there are no statistical param-
eters of the biosignal.
0 200 400 600 800 1 000 12 00 1 400 16 00 180 0
40
30
20
10
0
10
20
0 200 400 6 00 800 10 00 1 200 140 0 1600
40
30
20
10
0
10
20
180 0
subsampling
1800 samples
180 samples
Time(ms)
Time(ms)
mean
maximum
minimum
standard error
AmplitudeAmplitude
Figure 2: Illustration of the effect produced by a subsam-
pling operation over a random signal (adimensional ampli-
tude).
LONG TERM BIOSIGNALS VISUALIZATION AND PROCESSING
403

z =
log(N)
log(r)
−
log(npviz)
log(r)
+ 1
(2)
The correct zoom level, z, corresponding to each
selected zoom window is obtained with equation 2,
where N is the number of points that we are trying to
see and npviz is the maximum number of points to be
displayed (npviz and r are specified by the user). dxe
represents the ceiling operation (rounding for the next
integer).
2.3 Long Term Biosignals Processing
Besides visualization problems, long term biosignals
also need different processing approaches. Since we
are working with very large datasets, the processing
algorithms’ input can’t be the entire signal. The im-
plemented processing algorithms map the signal in in-
tervals with fixed length and process each mapped in-
terval, using an algorithm that works efficiently with
shorter signals. After processing each interval, the re-
sults are merged together.
X = {x
1
, x
2
, . . . , x
k
} (3)
Y = F(X ) (4)
Hereafter, we consider the discrete biosignal to be
processed, X, described in equation 3, where k repre-
sents the signal’s number of samples. The processing
operation can be represented by equation 4. The op-
erator F receives an entire biosignal (X) as input and
returns Y . Since the input signal might be very long,
X can be splitted in subgroups with a fixed number
of samples - L. The signal mapper is a list of pairs
that define the several subgroups to be processed sep-
arately (see J in equation 5).
J = {(0, L), (L − v, L − v + L),
(2L − 2v, 2L − 2v + L),
. . . ,
(mL − mv, mL − mv + L)} (5)
v is the number of samples to be overlapped, and
m an integer. Selecting the signal (X ) in the time in-
tervals defined by J, the signal will be mapped. Each
part of the signal can be defined by equation 6.
x
0
= {x
0
, . . . , x
L
}
x
1
= {x
L−v
, . . . , x
L−v+L
}
··· (6)
In order to avoid problems in the borders where
the signal is splitted, the implemented algorithm has
an overlapping number of samples, v (every time the
algorithm runs for a selected time window, there is a
number of samples from the end of the last time win-
dow that is in tthe beginning of the actual one). After
mapping the signal, the processing algorithm is ap-
plied to the various intervals mapped from the signal
and a group of outputs is obtained (equation 7).
y
j
= f (x
j
) (7)
On this last step, in which j represents a subpro-
cessing group, the results from the independent pro-
cessing tasks are merged together. The function that
correctly joins together the outputs from the subpro-
cessing tasks is denoted by G. The final result is given
by equation 8.
Y = G(y
0
, y
1
, . . . ) (8)
A mature algorithm (Pan and Tompkins, 1985),
which do not work properly on long term biosignals
was adapted: the ECG peak detector.
Considering a processing operation with a fixed
start time (T
s
), that takes a time T to be carried out by
one processor and that the processing is going to be
divided by N
s
processors, the total parallel processing
time (T
p
) will be given by equation 9 (the multiplica-
tion by (1 + Ov) avoids the result to be zero).
T
p
= T
s
+
T
N
s
× (1 + Ov) (9)
O
v
=
v
N
slice
(10)
The overlap (Ov) is defined by equation 10, where
N
slice
is the number of samples of each processing
slice. Since the existence of the overlap means that
there are samples being processed in two different
subtasks, a bigger overlap causes the processing to
last longer. However, if the overlap is too small, there
is the danger of occurring processing errors. In order
to prevent these errors, our ECG peak detection al-
gorithm only considers the data to be efficiently pro-
cessed when there are coincident peaks in the output
(adjacent subtasks detect at least one common peak).
3 PERFORMANCE EVALUATION
Several types of biosignals such as as electromyog-
raphy, electrocardiography, electrodermal activiy, ac-
celerometer and respiration were acquired in order to
test the visualization and processing algorithms. The
acquisitions were carried out at the patients’ homes,
with their approval, during the night (each recording
had the approximate duration of 8 hours), using a bio-
PLUX research system, a wireless signal acquisition
unit (PLUX, 2011).
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
404
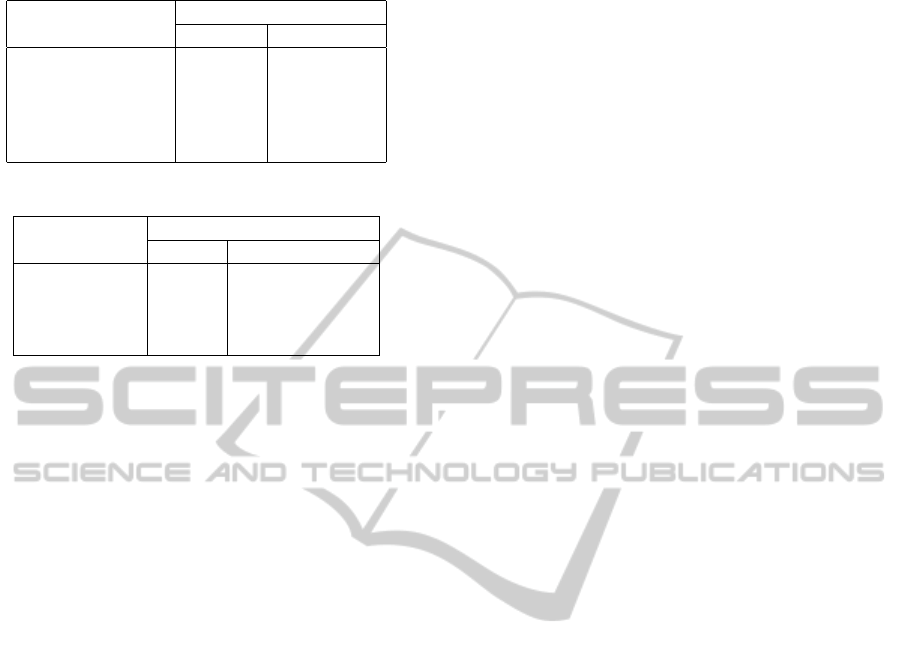
Table 1: Conversion times.
text file size (MB)
Conversion times (s)
raw data zoom levels
346,8 41 85
435,1 50 104
954,6 91 217
1.021,2 109 234
1.297,3 157 357
Table 2: Load times for .txt and .h5 files
file size (MB)
Load times (s)
.h5 file .txt file
14 0.01 6.35
144 0.04 64.33
347 0.57 349.33
424 0.79 (Memory Error)
3.1 Results and Discussion
All the performance tests were made with a Intel Core
i7 720QM laptop, with 1.60GHz processor. Regard-
ing data conversion to the new data structure, the per-
formance results are described in table 1.
Considering that opening text files with huge sizes
by loading them on python would take a long time
or even cause a memory error, the results presented
in table 2 evidence the benefits of the developed data
structure on data accessing. The performance of the
visualization tool is independent of the type and size
of the signal being visualized as well as of the zoom
level on which the user is ”navigating” with the de-
veloped tool. Operations like zooming and panning
over long term biosignals, that take several seconds
using python visualization methods, are almost in-
stantaneous using the developed tools. Since the data
structure creation only has to be carried out once, en-
abling instant accessing to data, it is possible to un-
derstand the advantages of the presented tools.
4 CONCLUSIONS AND FUTURE
WORK
Considering standard formats for biological and phys-
ical signals, it is easy to see that the developed data
structure allows a broader approach to the visual-
ization and processing of biosignals (particularly for
long term biosignals). Besides allowing the user to
save the raw data from the acquisition and important
information about the subject or the recorded signals
and the results of the parallel biosignal processing al-
gorithms. This format allows a new way of explor-
ing biological data, in a fast and intuitive multi-level
visualization of the biosignals. Since the developed
visualization tools are compatible with the web envi-
ronment, they can be used in the Internet.
In future work we aim to create an algorithm that
allows processed data visualization, as a way to link
the processed data and the signal, making it possible
to visualize at the same time the signal and important
processed data. Other future goal is to develop new
processing algorithms adapted to long term biosig-
nals, such as the heart rate variability, since it’s pa-
rameters are of great importance in clinical cases that
need long term monitoring. Regarding parallel pro-
cessing techniques, some improvements are still nec-
essary, such as an automatic calculation of the indi-
cated number of overlapping samples.
ACKNOWLEDGEMENTS
This work was partially supported by National
Strategic Reference Framework (NSRF-QREN) un-
der project ”LUL”, ”wiCardioResp” and ”Do-IT”,
and Seventh Framework Programme (FP7) program
under project ICT4Depression, whose support the au-
thors gratefully acknowledge. The authors also thank
PLUX, Wireless Biosignals for providing the acquisi-
tion system and sensors necessary to this work.
REFERENCES
HDF group (2007). HDF5 - HDF group [online] Avail-
able at:http://www.hdfgroup.org/HDF5/ [Accessed 5
September 2011].
Kayyali, H., Weimer, S., Frederick, C., Martin, C., Basa,
D., Juguilon, J., and Jugilioni, F. (2008). Remotely
attended home monitoring of sleep disorders. Telemed
J E Health, 14(4):371–4.
Pan, J. and Tompkins, W. (1985). A real-time qrs detection
algorithm. Biomedical Engineering, IEEE Transac-
tions on, (3):230–236.
Pinto, A., Almeida, J. P., Pinto, S., Pereira, J., Oliveira,
A. G., and de Carvalho, M. (2010). Home telemoni-
toring of non-invasive ventilation decreases healthcare
utilisation in a prospective controlled trial of patients
with amyotrophic lateral sclerosis. Journal of Neurol-
ogy, Neurosurgery & Psychiatry.
PLUX (2011). PLUX - Wireless Biosignals [online]
Available at: http://plux.info/ [Accessed 5 September
2011].
Sousa, J., Palma, S., Silva, H., and Gamboa, H. (2010).
aal@ home: a new home care wireless biosignal mon-
itoring tool for ambient assisted living.
LONG TERM BIOSIGNALS VISUALIZATION AND PROCESSING
405
