
BRAIN SEGMENTATION IN HEAD CT IMAGES
Ana Sofia Torres and Fernando C. Monteiro
Polytechnic Institute of Braganc¸a, Campus Santa Apol´onia, Apartado 1134, 5301-857 Braganc¸a, Portugal
Keywords:
Brain segmentation, Graph clustering, Head CT images, Watershed transform.
Abstract:
Brain segmentation in head computed tomography scans is essential for the development of computer-aided
diagnostic methods for identifying the brain diseases. In this paper we present a hybrid framework to brain
segmentation which joints region-based information based on watershed transform with clustering techniques.
A pre-processing step is used to reduce the spatial resolution without losing important image information. An
initial partitioning of the image into primitive regions is set by applying a rainfalling watershed algorithm on
the image gradient magnitude. This initial partition is the input to a computationally efficient region segmenta-
tion process which produces the final segmentation. We have applied our approach on several head CT images
and the results reveal the robustness and accuracy of this method.
1 INTRODUCTION
Image segmentation is one of the largest domains in
image analysis, and aims at identifying regions that
have a specific meaning within images. The role
of imaging as complementary mean of diagnosis has
been expanding beyond the techniques of visualiza-
tion and checkups in anatomical structures. This area
has become a very useful tool in planning of surgical
simulations and location of pathologies.
The Computed Tomography (CT) is an imaging
modality that allows the imaging of sections of the
human body, with almost no overlap of organs or
anatomical structures. Thus allowing us to actually
doing tests with a large number of sections quickly
and with high spatial resolution. The need for quanti-
tative analysis in tests with many sections has served
as a stimulus for the development of computational
methods for the detection, identification and delin-
eation of anatomical structures. The segmentation of
the brain from CT scans is an important step before
the analysis of the brain. This analysis can be per-
formed by a specialist, which manually surrounds the
area of interest on each slice of the examination. This
requires very careful and attentive work and practi-
cal exams with a high number of slices, the identifica-
tion of regions becomes a tedious and time consuming
task, subject to variability depending on the analyzer,
which makes it desirable to have automated methods.
However, if on one hand, manual segmentation has
the problems mentioned above, the automatic identifi-
cation of structures from CT images becomes a tricky
task not only because of the volume of data associ-
ated with the imaging study, but also the complexity
and variability in the anatomical study, and that noisy
images can provide. So developing new accurate al-
gorithms with no human interaction to segment the
brain precisely is important.
The watershed algorithm is an example of a hy-
brid method, combining information about the inten-
sity and the image gradient. This algorithm is a pow-
erful edge-based method of segmentation, developed
within the framework of mathematical morphology
(Vincent and Soille, 1991; Grau et al., 2004). Some-
times, the use of the watershed over-segmentation re-
sults in unwanted regions. To circumvent this prob-
lem markers are applied to the image gradient in order
to avoid over-segmentation, thus abandoning the con-
ventional watershed algorithm (Shojaii et al., 2005).
This operation allows the reduction of regional min-
ima, grouping them in the region of interest.
The proposed methodology in this paper has three
major stages. First, from the gradient image we
create, based on the watershed transform, an over-
segmented image. The regions formed are atomic
regions. In the next step, the region similarity
graph (RSG) will be created (Monteiro and Campilho,
2008), from the over-segmented image, for apply a
graph clustering approach in the last station. This
framework integrates edges and region-based seg-
mentation with spectral based clustering through the
watershed transform. Figure 1 presents the stages of
434
Sofia Torres A. and C. Monteiro F..
BRAIN SEGMENTATION IN HEAD CT IMAGES.
DOI: 10.5220/0003794704340437
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2012), pages 434-437
ISBN: 978-989-8425-89-8
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)
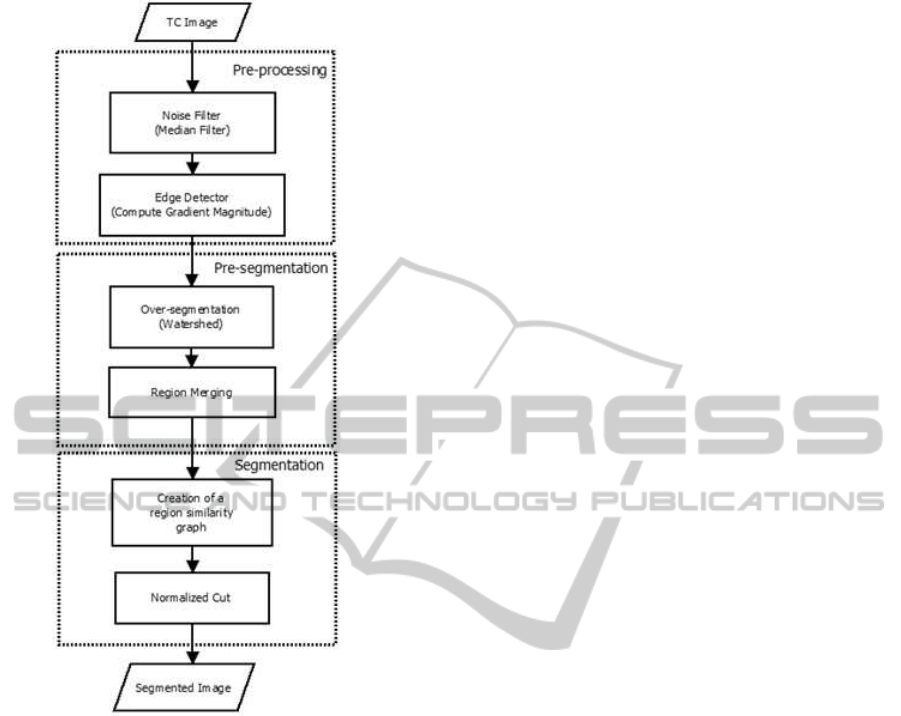
Figure 1: Phases of the proposed method.
the entire process.
The combination of watershed and clustering
methods solves the weaknesses of each method.
Rather than clustering single feature points we will
cluster small regions, confident that the underlying
primitive regions are reliable. Our approach actu-
ally prefers the objects to be over-segmented into a
number of smaller regions to ensure that a minimal
amount of backgroundis connected to anyof the brain
regions.
The algorithm described in this paper can be well
classified into the category of hybrid techniques, since
it combines the edge-based, region-based and the
morphological techniques together through the spec-
tral based clustering approach. We propose that our
method can be considered as an image segmentation
framework within which existing image segmenta-
tion algorithms that produce over-segmentation may
be used in the preliminary segmentation step.
The remainder of this paper is organized as fol-
lows. Section 2 gives a description of the methods
used in this paper. Followed by the experimental re-
sults and discussion in Section 3. The concluding re-
marks are given in the last section.
2 BRAIN SEGMENTATION
The segmentation of an image is one of the most im-
portant factors in the analysis and identification of the
brain on CT images. One of the objectives of devel-
oping new algorithms for image segmentation is to
increase the accuracy by reducing the computational
cost (Pham et al., 2000).
The watershed transform partitions the image in to
numerous regions depending on the number of local
minima of the gradient, usually the watershed tends
to produce an over-segmentation(Callaghan and Bull,
2005). In order to facilitate the calculations are over-
segmentation can be eliminated by incorporating a
pre-processing of the image. Many methods have
been proposed in order to reduce unwanted regions
and produce a meaningful segmentation.
In this work are provide some methods to over-
come this problem. For example, if the images con-
taining noise the first step is to use non-linear filters
such as the bilateral anisotropic filter, which smooths
images while preserving its contours and structure,
since it only acts on neighbours who are part of the
same core region. The next step is to eliminate the
weaker contours through gradient minima suppres-
sion, the process known pre-flooding (See Fig. 2).
This methodology uses a constant depth of certain
basin. Before to the transform each catchment basin is
flooded up to a certain height above its bottom, this is,
the lowest gradient magnitude and it can be thought
as a flooding of the topographic image at a certain
level (flooding level). In the latest step, is made by a
segmentation fusion, merging atomic regions with a
graph-based clustering approach.
Segmentation result directly using watershed al-
gorithm is shown in Fig. 2.(a) and from it we can find
that serious over-segmentation (1587 atomic regions)
makes the result meaningless even when we use pre-
flooding as showed in Fig. 2.(b) and (c), with 1281
regions and 814 regions, respectively.
Spectral-based methods use the eigenvectors and
eigenvalues of a matrix derived from the pairwise
similarities of features (pixels or regions). This effect
is achieved by constructing a fully connected graph.
Considering all pairwise pixel relations in an image
may be too computational expensive. Unlike other
well known clustering methods (Shi and Malik, 2000)
which use down-sampling pixel-based to construct
the graph, our method is based on selecting links from
BRAIN SEGMENTATION IN HEAD CT IMAGES
435
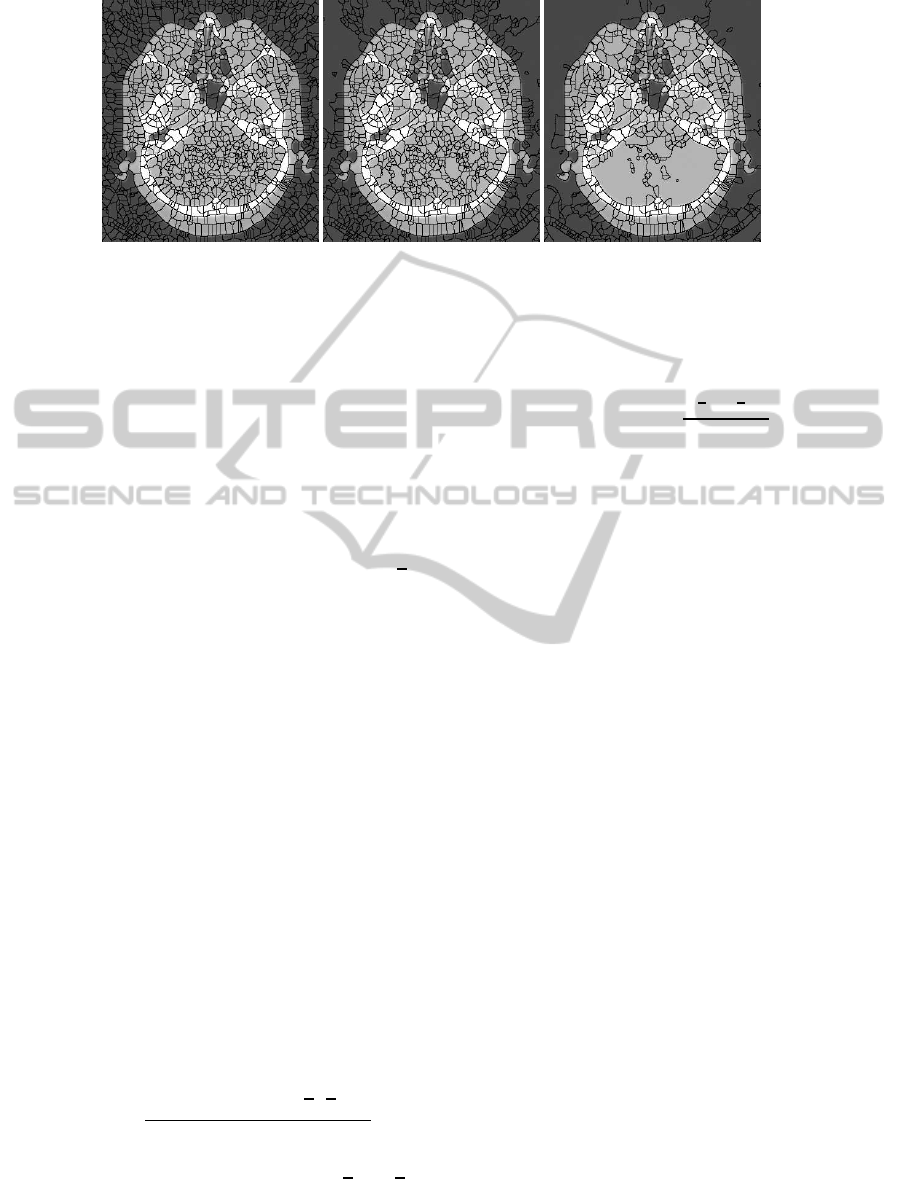
(a) (b) (c)
Figure 2: Atomic regions from watershed. (a) No pre-flooding. (b) Low level of pre-flooding. (c) Medium level of pre-
flooding.
a weighted undirected graph G = (V, E, W) based on a
region similarity graph where each node corresponds
to an atomic region (Monteiro and Campilho, 2008).
The proposed region similarity graph structure
takes advantage on region-based representation. The
set of nodes V are represented by the centroid of each
atomic region. The sets of links E and link weights
W represent, respectively, relationships and similarity
measures between pair of regions. Each region r
i
rep-
resents a small group of pixels where the centroid
x
i
is utilized as a node of the graph.
In almost all the graph-basedapproaches proposed
in the literature the spatial distance cue is also used to
compute the similarity between graph nodes. How-
ever, during our experiments, we noted that such cue
is responsible for the partition of homogeneous areas
in the image - an issue commonly associated to nor-
malized cut algorithm. Instead, we use intervening
contours (Leung and Malik, 2000) which are equiv-
alent to spatial distance without suffering from the
same problems. For each pair of nodes, node simi-
larity is inversely correlated with the maximum con-
tour energy encountered along the line connecting the
centroids of the regions. If there are strong contours
along a line connecting two centroids, these atomic
regions probably belong to different segments and
should be labeled as dissimilar.
Let i and j be two atomic regions and the orienta-
tion energy OE
∗
between them, then the intervening
contours contribution to the link weight is given by:
w
ic
(i, j) = exp
"
−
max
t∈line(i, j)
OE
∗
(
x
i
, x
j
)
2
σ
2
ic
#
,
(1)
where line(i, j) is the line between centroids
x
i
and x
j
formed by t pixels.
The mean intensity of each node contributes for
the link weight according to the following function:
w
I
(i, j) = exp
−
I
x
i
− I
x
j
2
σ
2
I
!
. (2)
These cues are combined in a final link weight
similarity function, with the values σ
ic
and σ
I
selected
in order to maximize the dynamic range of W:
W(i, j) = w
ic
(i, j) · w
I
(i, j) . (3)
To compute the similarity matrix the current ap-
proach uses only image brightness and magnitude
gradient. Additional features such as texture, could
be added to the similarity criterion. This may slow
the construction of the RSG but the rest of the algo-
rithm will proceed with no change.
3 EXPERIMENTAL RESULTS
The brain CT images used in this paper were pro-
viding by the database of IPB. The number of slices
varies from exam to exam, but not all slices of the
exam contain information regarding the brain, and for
that reason we used only a few images of a single scan
to his head. The images obtained in this condition are
stored with size of 512512 pixels. Each pixel is 16
bits in size and 0.85 mm resolution. The images are in
greyscale mode, stored in DICOM format. This group
of images selected includes the entire anatomy of the
brain from the top, the middle and the bottom. Figure
3 shows four slices selected from different parts of the
head to show the accuracy of this technique.
The brain segmentation results for the slices in
Fig. 3 are shown in Fig. 4. Comparing the segmented
brain regions with the original image confirms that
our approach separates accurately the brain regions.
BIOSIGNALS 2012 - International Conference on Bio-inspired Systems and Signal Processing
436
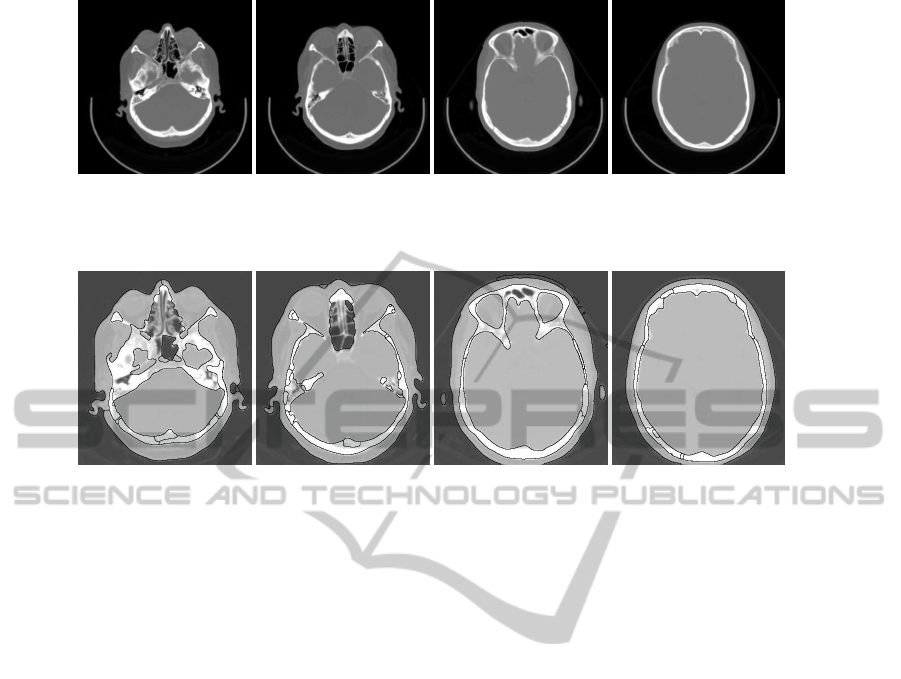
(a) (b) (c) (d)
Figure 3: Original CT head images.
(a) (b) (c) (d)
Figure 4: Brain segmented images corresponding to the slices in Fig. 3, respectively.
4 CONCLUSIONS
In this paper we have proposed an image segmenta-
tion methodwhich combines edge- and region-based
information with spectral techniques through the mor-
phological algorithm of watersheds. An initial parti-
tioning of the image into primitive regions is set by
applying a watershed simulation on the image gradi-
ent magnitude. This initial partition is the input to a
computationally efficient graph partition process that
produces the final segmentation. The latter process
uses a region similarity graph representation of the
image regions.
Using small atomic regions instead of pixels leads
to a more natural image representation - the pixels
are merely the result of the digital image discretiza-
tion process and they do not occur in the real world.
Besides producing smoother segmentations than pixel
based partitioning methods, it also reduces the com-
putational cost in several orders of magnitude.
As future work it would be interesting to obtain
brain contours in the region carried out by experts, to
be subsequently developed a methodology for evalu-
ating the accuracyof brain contours resulting from the
segmentation algorithms. This assessment method-
ology can understand some criteria to overcome the
subjectivity barrier between the silhouette of the brain
performed by different specialists.
REFERENCES
Callaghan, R. O. and Bull, D. (2005). Combined
morphological-spectral unsupervised image segmen-
tation. IEEE Trans. Image Processing, 14(1):49–62.
Grau, V., Mewes, A., Alcaniz, M., Kikinis, R., and Warfield,
S. (2004). Improved watershed transform for medi-
cal image segmentation using prior information. IEEE
Transactions on Medical Imaging, 23(4):447 –458.
Leung, T. and Malik, J. (2000). Contour continuity in
region-based image segmentation. In Procedings Eu-
ropean Conference in Computer Vision, volume I,
pages 544–549, Freiburg, Germany.
Monteiro, F. and Campilho, A. (2008). Watershed frame-
work to region-based image segmentation. In Pro-
ceedings of the International Conference on Pattern
Recognition, pages 1–4, Tampa, USA.
Pham, D., Xu, C., and Prince, J. (2000). A survey of cur-
rent methods in medical image segmentation. Annual
Review of Biomedical Engineering, 2:315–337.
Shi, J. and Malik, J. (2000). Normalized cuts and image
segmentation. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 22(8):888–905.
Shojaii, R., Alirezaie, J., and Babyn, P. (2005). Auto-
matic lung segmentation using watershed transform.
In Proc. of the International Conference of Image Pro-
cessing, volume II, pages 1270–1273, Genova, Italy.
Vincent, L. and Soille, P. (1991). Watersheds in digital
spaces: an efficient algorithm based on immersion
simulations. IEEE Transactions on Pattern Analysis
and Machine Intelligence, 13(6):583–589.
BRAIN SEGMENTATION IN HEAD CT IMAGES
437
