
UNIFIED PARALLEL EXPERIMENT INTERFACE FOR MEDICAL
RESEARCH SYSTEM
Michal Kratochv´ıl, Petr Vcel´ak and Jana Kleckov´a
Department of Computer Science and Engineering, University of West Bohemia, Univerzitni 8, Plzen, Czech Republic
Keywords:
Medical research system, ITK, Parallel processing, Interoperability.
Abstract:
When we are processing large quantity of a data, and/or we are computing complex tasks, computational
performance of one computer is not enough. Solution is parallel processing. However proper approach to
parallel programming doesn’t need to well-known to medical experts or computational tool doesn’t have native
support for parallel programming. Our goal is to design unified interface, which allows parallel approach to
our medical researchers. It must provide support for existing medical experiments and it must provide full
interoperability.
1 INTRODUCTION
The aim of this article is demonstration of unified
parallel interface for implementation medical exper-
iments. General design of our research information
system will be presented in (Vcelak and Kleckova,
2011). There is some interoperability issues between
experiments, which we need to solve during develop-
ment. Original design of research system supposed
running experiments as executable tasks. Our main
ideas were that each task must implement. Exper-
imental Application programming interface (EAPI)
and experiments are planned by Experiment Execu-
tion Planner (EPP). When we execute large amount
of experiments on large medical data, we have to deal
with following problems.
• Users (medical doctors) requested different tasks
priority, than experiment composers (program-
mers), e.g. they awaited examination result as-
sessments in short time (optimal in real-time),
while programmers assumed execution in night
time.
• There are development teams from different de-
partments. Each of them is using different pro-
gram tools, e.g. C, C++, C#, Perl, .NET, OWL
(Bodenreider and Stevens, 2006) and Matlab.
• There are different visualisation methods in use.
• An additional libraries e.g. .NET, or frameworks
e.g. ITK/VTK (Ib´a˜nez L., Schroeder W., Ng L.,
Cates J., 2003), MITK (Wolf et al., 2005) are re-
quired. Some of them are not open source (which
we prefer).
• There are experiments, which need large value of
RAM, or they consume large CPU time e.g. huge
matrix operations.
• It is hard to maintain all libraries and programs
without software conflicts.
• Dedicated machine is overloaded frequently..
• Some programming tools doesn’t native support
for working with medical data – DICOM (Na-
tional Institute of Neurological Disorders and
Stroke, 2010) and/or HL7 (Health Level Seven,
Inc., 2010) files.
We need to adapt current interface EAPI to pro-
vide fully interoperability between experiments
and programming languages. There are extra re-
quests to new EAPI.
• We required to provided functions for Parallel
computations.
• New EAPI have to be able to split up computa-
tions to single dedicated machines.
• Each of computation has to be able processed on
whatever machine, which is registered in research
system.
We designed extension to EAPI – Parallel Exper-
iment API (PEAPI), which is able to provide new
possibilities. Programmers are now able to use
parallel computation without special parallel pro-
gramming skills. The new PEAPI fulfilled these
requests, the following properties are achieved.
449
Kratochvíl M., Vcelák P. and Klecková J..
UNIFIED PARALLEL EXPERIMENT INTERFACE FOR MEDICAL RESEARCH SYSTEM.
DOI: 10.5220/0003873404490452
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2012), pages 449-452
ISBN: 978-989-8425-88-1
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)
• Central system not deal requests itself now.
• Central system distribute incoming requests to
single computers or computer centres/clouds.
2 PEAPI INTERFACE
DESCRIPTION
The new design is based on current design, so the hi-
erarchy of EAPI was preserved. There are new layers,
which provide functionality for new computer reg-
istration and a data distribution between them. The
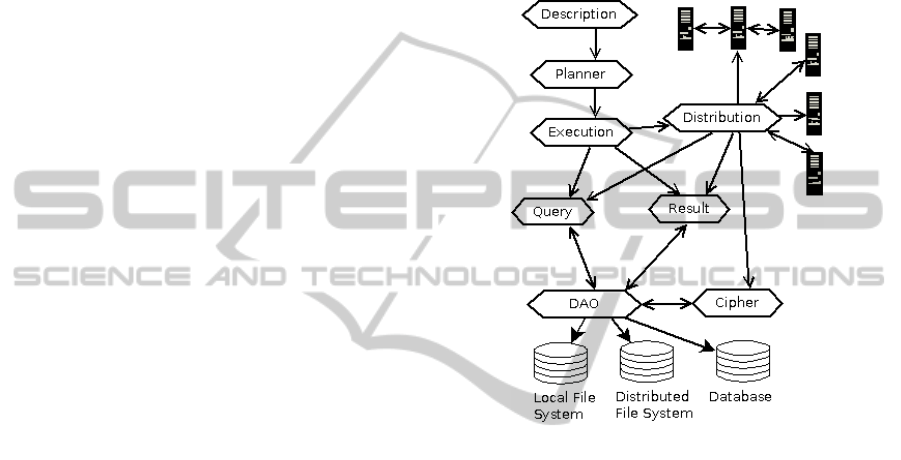
PEAPI hierarchy is shown at Figure 1.
Planner Layer. There are new properties, which al-
low experiments priority planning based on avail-
able resources in computing network and on-
demand data processing with highest priority
(needed for real-time results).
Execution Layer. This layer cooperates with new
distribution layer. Now it selects what type of
computation is used. This layer also provides
computation assigning to single computer, which
comply computation requests (when no parallel
computation is used).
Distribution Layer. It is a new layer. It tests avail-
able availability of computing machines with
proper programming languages, tools and/or
frameworks. It selects suitable parallelisation type
(depends on experiment code) and sends data to
a cipher layer, or DAO request to client comput-
ers. Distribution layer also includes planning al-
gorithms for node load and/or performance as-
sessment. Distribution layer is load-balancing
data to maximise system performance. This load-
balancer is based on current load and computer
performance indexes e.g. in the computing net-
work with many multi-processors and one single-
processor computer distribution layer assign only
small part of computation to single-processor
computer.
Cipher Layer. Most of the data are anonymised. But
their still has personal status. We need to secure a
data transfer through LAN or internet. We encrypt
only selected data e.g. DICOM or HL7 files or
VTK matrixes. We send raw or partial data not
encrypted, because we try to lower CPU load. The
data transfer still using DAO layer.
Query and Result Layers. Function of these layers
remain untouched. There are some new prop-
erties for composing partial results for/from net-
work nodes only. These layers were also ex-
tended with new functions to provide interoper-
ability with tools without native support of DI-
COM and/or HL7 files. It is often needed to dis-
tribute whole DICOM file set, but programs ex-
tracts only Meta data, or image data for further use
during computation. These main properties were
added to Query and Result layer which transfer
data as 3D matrix and meta-data objects. So we
can use DICOM/HL7 data in whole set of pro-
gram tools.
Figure 1: Description of new research system with parallel
support.
3 COMPUTING NETWORK
One of main problem, which we need to be solved,
was strongly heterogeneous programming language
environment. For example: we need to run liver seg-
mentation, followed-up by vein system reconstruction
and blood flowing rate computation. For these we
need 3 experiments. Each of them is using differ-
ent programming language (different research depart-
ments) – .NET, C++ with ITK/VTK and Matlab. We
designed the computing network (CN) to solve this
problem. The CN observe following rules.
• Experiments are assigned by central node (Client-
Server topology).
• The data distribution are managed by DAO layer,
which can be distributed to several computers.
• The computations are using master-slave or peer-
to-peer topology. Computers processing equal
data are using master-slave topology, but unde-
pendable nodes are using peer-to-peer connection,
when needed.
HEALTHINF 2012 - International Conference on Health Informatics
450
• We are using PVM (Oak Ridge National Labo-
ratory, 2011) and MPI (ANL Mathematics and
Computer Science, 2011) standards for parallel
computations.
We are using dedicated research system server
with database and WWW server to running central
node on it. Users can register their own computer, or
any other computer to the system. These computers
must have some of needed environment installed and
configured on it. The system executes compatibility
and performance tests first and creates performance
indexes and set of suitable computers, witch is shown
on Figure 2. After that, the distribution layer is able to
use these computers for computation, when it’s avail-
able.
4 PARALLEL PROCESSING
WITH PEAPI
There are several cases, we must deal with.
• Single Data, Multiple Processes (SDMP). Typ-
ically used, when we need parallelisation on ex-
periment level.
• Multiple Data, Multiple Process (MDMP). Typ-
ically used, when we need to process large data
quantity repeatedly and when we need to compare
single iteration results.
• Multiple Experiment, Multiple Process
(MEMP). Used, when we need repetition of
experiments (calculation faults, pre-processing or
partial computations, sets of computations).
The PEAPI allows programmers to call PEAPI
methods in their code. After that, PEAPI handles
process and data distribution, sub-process communi-
cation and parallel computation itself. There are not
extra demands on programmers. Programmers mark
parts of code as ready for parallel computation. Sim-
ple example code is shown below:
Program test;
Some variable declarations;
...
PEAPI_run_experiment_MEMP(
experiment_descriptor, count, data)
{
MyData[] = PEAPI_read_DAO_MDMP(
requested_data, cryptedKey);
...
Some operations
...
//call SDMP function to existing array
For (int I = 0; I <= sizeOf(MyData); i++)
{
MyProcessedData[] =
PEAPI_do_SDMP(MyData[i], PEAPI_function)
}
//create own SDMP code
For (int I = 0; I <= sizeOf(MyData); i++)
{
//create integer monitor
PEAPI_doMyCode_SDMP(){
PEAPI_protected_int myNumber;
Some code
...
myNumber++;
...
}
}
};
5 RESULTS
The liver segmentation and liver vein reconstruction
from CT example was selected for demonstration.
Experiment was executed on set of 50 patient’s DI-
COM CT studies. Some patients has damaged liver
with tumors, etc. A result of experiment is VTK ma-
trix with 3D surface and vein tree for each patient.
First measurement was serial computation. Reference
computer configuration was Pentium i5-750, 16 GB
RAM DDR3, SSD HDD, Debian Linux 6.0.2, stan-
dard package C++ compiler, VTK/ITK version 3.20.
Second and third measurement used this computer as
main node. CN consist of different 15 PC. Each of
them has 2 – 8 x processors (from old Pentium III
servers to new 6-core Xeon processors, with 1 – 8GB
RAM) CN is connected together by 100 Mbit Eth-
ernet. Second run used MEMP, third MDMP, fourth
MEMP + MDMP. Each measurement was executed 5
times, and averaged. Results are shown in the Table
1.
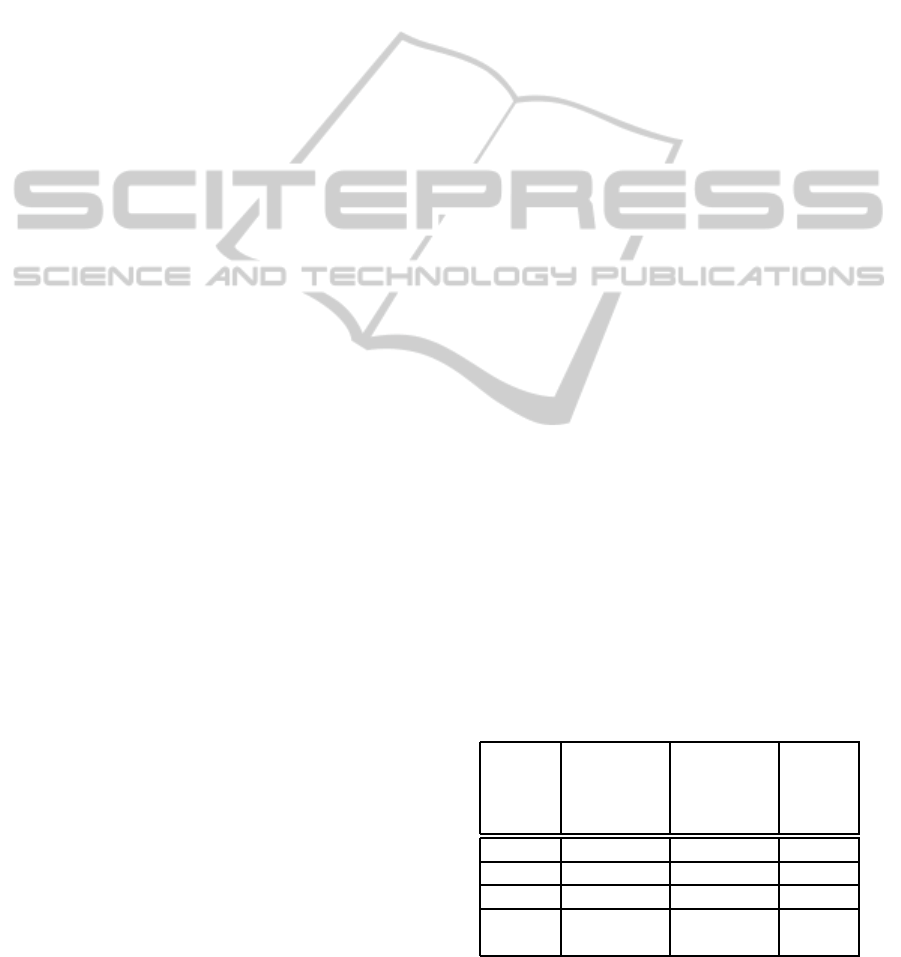
Table 1: Performance test: serial, MEMP and MDMP ap-
proaches.
Test Fastest
compu-
tation
[s]
Slowest
compu-
tation
[s]
All
jobs
done
[s]
Serial 140.2 145.1 7250.8
MEMP 98.3 358.3 479.1
MDMP 99.1 196.3 230.2
MEMP
MDMP
98.1 355.2 410.6
UNIFIED PARALLEL EXPERIMENT INTERFACE FOR MEDICAL RESEARCH SYSTEM
451
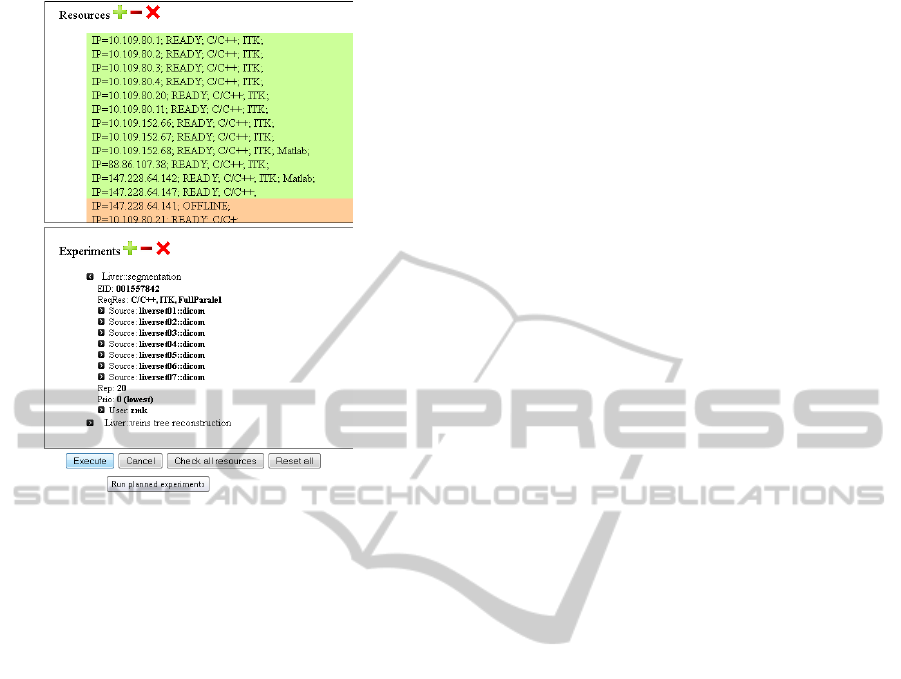
Figure 2: Experiment planning and resource checking
screen.
6 CONCLUSIONS
As is shown in Table 1, we can see large perfor-
mance increase after MEMP and/or MDMP is used.
Because we used real environment, we have hetero-
geneous node configuration. This is reason why in
MEMP the shortest and the longest time values dif-
fer so much (It means time computed on slowest and
fastest computer in network). We can also see that us-
ing both methods givesus similar results. It is because
of slowest computer on network. It is still working on
its data, while other computers are already done. In
worse case (which did not arise), the slowest com-
puter will start compute new task short moment be-
fore other finished. MDMP is shown as the best ap-
proach. It’s because the fastest computers do the most
jobs. The load-balancing is the most effective.
Thanks to PEAPI, we can process time-
demanding experiments. Distribution of environment
from one computer to many gives us better control on
each computer system and whole research system.
7 FUTURE WORK
Our future work is oriented to create PEAPI for other
programming languages, such as Matlab (now it’s
available for C/C++, Perl, .NET and other languages,
which supports PVM or MPI). Now we can only use
MEMP approach for others. We are also planning
connection to meta-centre for further computations,
which give us access to hundreds of computers.
ACKNOWLEDGEMENTS
The work presented in this paper was supported
by the project Czech Science Foundation number
106/09/0740.
REFERENCES
ANL Mathematics and Computer Science (2011). Message
passing interface (mpi) standard. Online, 2011-10-02.
http://www.mcs.anl.gov/research/projects/mpi/.
Bodenreider, O. and Stevens, R. (2006). Bio-ontologies:
current trends and future directions. Briefings in
Bioinformatics, 7(3):256 – 274.
Health Level Seven, Inc. (2010). What is hl7? Online,
2011-10-02. http://www.hl7.org/about/index.cfm.
Ib´a˜nez L., Schroeder W., Ng L., Cates J. (2003). The ITK
Software Guide, Kitware.
National Institute of Neurological Disorders and Stroke
(2010). Digital Imaging and Communications
in Medicine (DICOM). Online, 2011-10-01.
http://medical.nema.org.
Oak Ridge National Laboratory (2011). Parallel
Virtual Machine (PVM). Online, 2011-10-02.
http://www.csm.ornl.gov/pvm/.
Vcelak, P. and Kleckova, J. (2011). Semantically inter-
operable research medical data and meta data extrac-
tion strategy. 2011 4rd International Conference on
Biomedical Engineering and Informatics BMEI 2011,
4:1950 – 1954.
Wolf, I., Vetter, M., Wegner, I., B¨ottger, T., Nolden, M.,
Sch¨obinger, M., Hastenteufel, M., Kunert, T., and
Meinzer, H.-P. (2005). The medical imaging interac-
tion toolkit. Medical Image Analysis, 9(6):594 – 604.
HEALTHINF 2012 - International Conference on Health Informatics
452
