
DISEArch
A Strategy for Searching Electronic Medical Health Records
David Elias Pe
˜
na Clavijo, Alexandra Pomares Quimbaya and Rafael A. Gonzalez
Departamento de Ingenier
´
ıa de Sistemas, Pontificia Universidad Javeriana, Bogot
´
a, Colombia
Keywords:
Medical Health Records, Health Data Mining, Text Mining.
Abstract:
This paper proposes DISEArch, a novel strategy for searching electronic health records (EHR) of patients
that have a specific disease. The objective of DISEArch is to enhance research activities on disease analysis
allowing researchers to describe the disease they are interested on, and providing them the EHRs that best
match their description. Its principle is to improve the precision of searching EHRs combining the analysis of
structured attributes with the analysis of narrative text attributes producing a semantic ranking of EHRs with
respect to a given disease. DISEArch is useful in medical systems where the information about the primary
diagnosis of patients may be hidden in narrative text hindering the automatic detection of relevant records for
clinical studies.
1 INTRODUCTION
Electronic health records (EHR) are a rich source of
knowledge for medical research. However, their use
has been limited due to the fact that important infor-
mation is stored in narrative texts, intended for hu-
mans, difficult to search and analyse automatically.
One of the requirements of medical research is to find
the EHRs of patients that have been diagnosed with
a specific disease. This task that should be easily
done using classical queries (e.g. SQL) is very time-
consuming because diagnosis is frequently hidden in
the text (e.g. medical notes), hindering the possibil-
ity of automatically detecting relevant records and re-
quiring the participation of an expert. Previous work
on EHR systems propose strategies to improve auto-
matic processing of narrative text in EHRs using in-
formation retrieval and data mining techniques (Han
et al., 2006)(Zhou et al., 2005).
This work proposes DISEArch, a strategy for search-
ing in EHRs those records that match a specific di-
agnosis, regardless of the kind of attribute (structured
or non structured) that contains the information. DIS-
EArch is composed of three phases. The first extracts
the set of patient records from the medical health sys-
tem. The second phase applies classical queries on
structured attributes and text mining techniques over
narrative text. Finally, it ranks the records by apply-
ing a semantic distance function with respect to the
given disease description. DISEArch has been useful
in reducing the time required for searching medical
records. The structure of the paper is as follows. Sec-
tion 2 presents the analysis of related works on nar-
rative text and medical record analysis. Section 3
presents DISEArch, including its main components.
Section 4 presents the main aspects of the prototype
of DISEArch and the evaluation of its behaviour. Fi-
nally, Section 5 concludes this paper.
2 RELATED WORKS
Figure 1 presents a taxonomy of existing works re-
lated to text mining from EHRs. The initial categories
offered are general approaches, algorithms, tools and
scope. General approaches refer to three main bod-
ies of work: information retrieval, natural language
processing (NLP) and text/data mining. Algorithms
are further divided into those aimed at data prepara-
tion and those aimed at data detection or classifica-
tion. Tools offers a list of some available software
tools which may support the process of text mining
from EHRs. Finally, scope centers on work aimed
at analyzing negated sentences, as opposed to work
which is more generic. In our taxonomy (Figure
1) general approaches start with text mining, which
consists of analyzing (portions of) documents typi-
cally made up of natural language. Its purpose is
to uncover patterns, trends and relationships between
words, meanings, terms or concepts (Spasic et al.,
151
Elias Peña Clavijo D., Pomares Quimbaya A. and A. Gonzalez R..
DISEArch - A Strategy for Searching Electronic Medical Health Records.
DOI: 10.5220/0004004601510156
In Proceedings of the 14th International Conference on Enterprise Information Systems (ICEIS-2012), pages 151-156
ISBN: 978-989-8565-10-5
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)
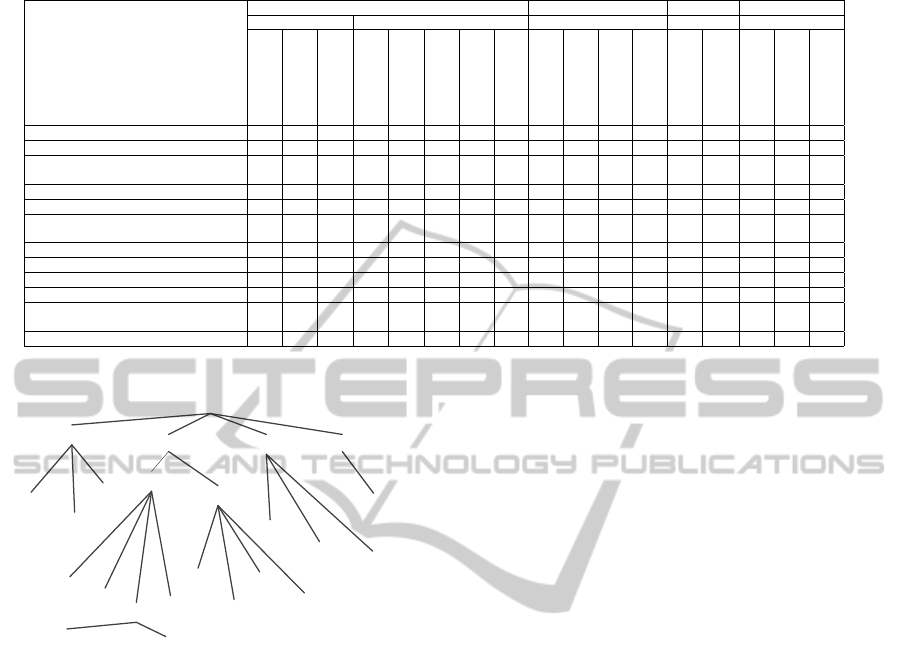
Table 1: Comparative literature review.
Algorithms Tools Approach Field
Preparation Classification/Detection
Papers
Data transform.
Expert Tagging
UMLS Tagging
Regexp
Proposed
Bayesian networks
Decision trees
Hidden Markov M.
GATE
Link G. Parser
EMERSE
Own
Negated sentences
Generic
NLP
Data/text mining
IR
NegEx (Chapman et al., 2001)
√ √ √ √ √ √
Context-Sensitive (Averbuch et al., 2004)
√ √ √ √ √ √ √ √
Negation-Recognition (Rokach et al.,
2008)
√ √ √ √ √ √ √ √ √
Geneneric Extraction (Han et al., 2006)
√ √ √ √ √ √
DM & CBR (Huang et al., 2007)
√ √ √ √ √ √ √
Text mining in biomedicine (Spasic et al.,
2005)
√ √ √ √ √
Diabetic DW (Breault et al., 2002)
√ √ √ √ √ √
EMERSE (Seyfried et al., 2009)
√ √ √ √
ABN (Antal et al., 2001)
√ √ √ √
Decision-Making (Claster et al., 2008)
√ √ √ √ √ √ √
Semi-structured data to knowledge (Zhou
et al., 2005)
√ √ √ √ √ √ √ √ √
HMM & LSA (Ginter et al., 2009)
√ √ √ √ √ √ √
DATA ENGINEERING TECHNIQUES FOR ELECTRONIC MEDICAL RECORDS
Algorithms
Tools
Approach
Field
Preparation
Classification /
Detection
Tokenization
Tagged
Lemmatization
Stemmer
Regular
Expresions
Decision
Trees
Bayesian
Networks
GATE
EMERSE
Link
Grammar
Parser
Negated
Sentences
Information
Retrieval
Text/Data
Mining
Natural
Processing
Language
Expert
UMLS
Hidden
Markov
Models
Figure 1: Taxonomy of EHRs data techniques.
2005). The second general approach is related to the
field of information retrieval (IR)(Manning et al.,
2008). The last general approach deemed useful for
our purposes is natural language processing (NLP),
which refers to the recognition and use of informa-
tion expressed in human language through computer-
based systems (Hotho et al., 2005).
Our taxonomy continues by classifying specific types
of algorithms that can be used as part of the three
general approaches, depending on the stage of the
process. With regards to data preparation we in-
clude four kinds of algorithms that prove useful in
preparing unstructured health records prior to analy-
sis.Tokenization is the process through which a flow
of text is divided into segments. Lemmatization
refers to a method in which verbs are transformed
into their base form or nouns into their singular form.
Stemming is used for removing irrelevant terms from
the text. The last type of algorithm is tagging, which
involves the interaction with a user that labels the text.
In the case of EHRs, these tags are typically part of
a controlled vocabulary, such as UMLS (USNLM,
2011). The second general type of algorithm in our
taxonomy is grouped under classification and de-
tection. Decision trees are a common part of the
tool-belt for data mining and are useful in classifying
conditions hierarchically such that a final decision is
reached when a path can be followed from the root to
one of its leaves. Bayesian networks are a powerful
tool which is implemented through acyclic directed
graphs that contain a set of nodes, each representing a
random variable. (Antal et al., 2001).
A related kind of algorithm is called hidden Markov
model, which represents a statistical model for lin-
ear problems and is widely used for speech recogni-
tion (Ginter et al., 2009). The third branch of related
works is focused on the tools that support data and
text mining in EHRs. Among these we find GATE
(General Architecture for text engineering) which of-
fers a general open source framework for develop-
ing or deploying software components for text en-
gineering (Cunningham et al., 2011). Another tool
is the Link Grammar Parser, which syntactically
analyses text based on link grammar. EMERSE (The
Electronic Medical Record Search Engine) (Hanauer,
2006) is specifically aimed at EHRs, acting as a
search engine for free text inside such records. Us-
ing the taxonomy proposed above, Table 1 presents a
comparative review of relevant literature around data
/ text mining in EHRs.
3 DISEArch STRATEGY
The strategies to examine narrative texts described in
Section 2, provide a broad knowledge base to address
the analysis of unstructured text inside EHRs. How-
ICEIS2012-14thInternationalConferenceonEnterpriseInformationSystems
152

ever, they are focused exclusively on the analysis of
narrative text without taking into account the depen-
dencies on other (structured) fields within the record.
This work explores the combination of structured and
narrative analysis to enhance the precision on the se-
lection of relevant records for medical research. The
strategy proposed in this section, called DISEArch,
allows researchers to describe the disease they are in-
terested in and provides them the set of health records
that better match their description.
3.1 Phases
The process of analyzing health records in DISEArch
is divided into the phases illustrated in Figure 2. In
Describe
Disease
Preselected
Records
Analyse
Structured
Attribute
Analyze
Narrative
Results
Integrate
Results
Calculate
Semantic
Distance
Disease
Preselected
Records
User
Disease
Knowledge base
Disease
Disease
Records
Disease
Enriched
Diesase
Disease
Match Result
Match Result
Prioritized Dealth
Records
Result
Disease Enriched
Enrich
Description
Figure 2: DISEArch Strategy.
the first phase DISEArch allows medical researchers
to describe the disease they are interested in using a
template. This template includes formal and informal
aspects of the disease, including the scientific name,
the informal name, the tests that are typically used to
diagnose the disease, and the symptoms of the dis-
ease. Once the disease is described, each of the fields
are enriched using knowledge about the disease. The
enrichment is made using a knowledge base created
in OWL (Bechhofer et al., 2009) using a MeSH based
thesaurus
1
.
The set of preliminary records are stored in a local
database where DISEArch executes the analysis. The
goal is to find the elements described in the disease
within each one of the selected records, regardless of
whether they are contained in an structured or a nar-
rative text field. After this analysis, a score is given to
each one and then prioritized.
3.2 Search Process
The disease description is divided into n subgroups S
that are composed by m literals L ( Definition 1). An
example of subgroup is Disease Name and its liter-
als are Scientific name, Formal name, Informal name,
Synonyms and Acronyms. Similarly, health records M
1
http://www.nlm.nih.gov/
are divided into a set of p structured attributes S and q
narrative text attributes T (Definition 2).
Definition 1 . Disease Description. A Disease def-
inition D is composed of a set of subgroups S =
{s
1
, s
2
, ..., s
n
} that describe the main characteristics
of the disease. Each subgroup s
i
is specialized in a
view of the disease and is composed of a set of liter-
als L(s
i
) = {l
i1
, l
i2
, ..., l
im
} where l
i j
represents a fixed
value for an atomic characteristic of the disease.
Definition 2 . Health Record. A health record M
is composed of a set of structured attributes C =
{c
1
, c
2
, ..., c
p
} whose domain of values is discrete and
a set of narrative text attributes T = {t
1
, t
2
, ..., t
q
}
whose domain is a natural language text.
The goal of the search process is to detect within C
k
and T
k
of a record M
k
, the value of each one of the
literals l
i j
. If the value of the literal l
i j
is found in at
least one attribute of the record M
k
the value of the
search process is changed to one (1), otherwise it is
left at zero (0).
DISEArch contains two search functions in charge of
detecting the occurrence of literal values into health
records; the first one detects the value of a literal in
structured attributes C and the second one searches
within narrative text attributes T . Searching struc-
tured attributes is straightforward using classic sql
queries. On the contrary, searching within narrative
texts includes a previous preparation of texts and anal-
ysis that is detailed in Algorithm 1.
Algorithm 1: Narrative text search function.
Require: Record narrative text attributes
Ensure: Record search result
1: i,j,result ⇐ 0
2: for all textAttribute in record do
3: p ⇐ prepareText(textAttribute)
4: for all subgroup in diseaseTemplate do
5: for all literal in subgroup do
6: ortResult ⇐ searchValue(p).
7: if ortResult = 1 then
8: semResult ⇐ searchContext(p)
9: if semResult = 1 then
10: result ⇐ 1
11: end if
12: end if
13: j ⇐ j + 1
14: end for
15: i ⇐ i + 1
16: end for
17: recordResult[i, j] ⇐ result
18: end for
19: return recordResult
DISEArch-AStrategyforSearchingElectronicMedicalHealthRecords
153

At the end of the search process the output is the
score for each literal as the matrix Res illustrates and
the number of hits for each one of the literals. The
columns of Res represent the literals of each subgroup
and the rows the health records.
Res =
l
11
l
12
| ··· |l
n1
l
n2
l
n3
M
1
1 1 ··· 1 1 1
M
2
0 0 ··· 1 1 0
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
M
k
0 1 ··· 1 0 0
M
r
1 0 ··· 0 0 0
3.3 Integration Process
The integration process is in charge of representing
the results of the search process taking into account
the semantics of subgroups. This integration includes
the results provided by structured and non-structured
search functions. For doing this new representation
the process takes into account the Definitions 3, 4, 5.
Definition 3 . Subgroup Intensity. The intensity I of
a subgroup of literals s
i
is the normalised percent-
age of matched literals within the health record M
j
.
If a literal has multiple possible values (e.g. multi-
ple acronyms) each value is considered a literal (e.g.
Acronym 1, Acronym 2, etc.).
Definition 4 . Subgroup Utility. The utility U of a
subgroup of literals s
i
is a percentage value of the im-
portance it has in identifying the disease diagnosed
in a health record assuming that all the values of the
literals are positive.
Definition 5 . Subgroup Level of Hits. The number
of hits H of a subgroup s
i
is the normalised number of
times that literal values were matched within M
j
.
In order to calculate the utility of each subgroup
DISEArch uses a classical method of multi-criteria
decision analysis where each subgroup is evaluated
on multiple criteria by experts and the utility is “the
average specified in terms of normalised weightings
for each criterion, as well as normalised scores for
all options relative to each of the criteria” (Keeney
and Raiffa, 1976). The number of hits is used as
an optional calibration value that takes into account
the number of times that literal values are found in
a health record. The intention is to assign a higher
weight to records that have the same literal multiple
times. This value is optional because for some sub-
groups it is important, but for others it is not. At
the end of the integration process an integration ma-
trix is generated (see Matrix I). The values of the lit-
eral in each subgroup are described in the following
columns:
1. s
i
is 1 if at least one of the literals of the subgroup
was found in the health record M
k
, otherwise its
value is 0.
2. s
I
i
is the intensity of the subgroup.
3. s
U
i
is the utility of the subgroup to detect the dis-
ease.
4. s
H
i
is the number of hits of the subgroup. This
columns is optional.
I =
s
1
s
I
1
s
U
1
s
H
1
| ··· |s
n
s
I
n
s
U
n
M
1
1 1 0.6 1 ··· 1 1 0.4
M
2
0 0 0.6 0 ··· 1 0.66 0.4
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
M
k
1 0.5 0.6 0.5 ··· 1 0.33 0.4
M
r
1 0.5 0.6 0.7 ··· 0 0 0.4
The distance function between the disease de-
scription D and each one of the analyzed health
records M is calculated using a distance function (e.g.
Euclidean, Manhattan). The disease description as
well as each record are represented in a n-space (see
Function 1 and 2, respectively), where n is the number
of subgroups.
D = (p
s
1
, p
s
2
, ..., p
s
n
). (1)
M = (q
s
1
, q
s
2
, ..., q
s
n
). (2)
The value of each point p is equivalent to s
U
i
and
the value of each point q is calculated using the prod-
uct of s
U
i
×s
I
i
×s
H
i
. The record with the shortest dis-
tance is the first one in the prioritized list and so on.
4 IMPLEMENTATION AND
VALIDATION
In order to evaluate DISEArch and validate its im-
provement on the selection of the most relevant health
records given a disease, a prototype has been con-
structed and used to evaluate its precision and recall.
This section presents the main results obtained during
this evaluation.
4.1 Prototype
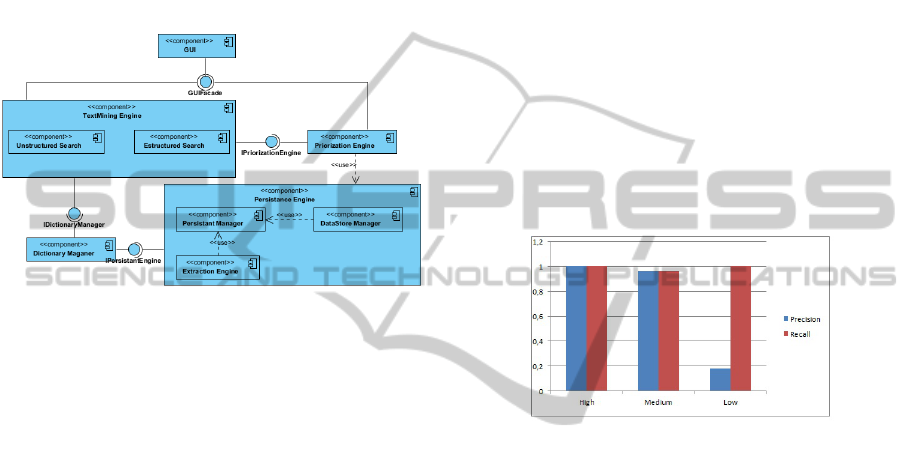
For evaluating the behaviour of DISEArch we devel-
oped the components presented in Figure 3. These
components are written in Java. The template of the
disease can be filled using the GUI or directly using
an XML file. The Dictionary Manager handles the
knowledge base that allows the enrichment of the de-
scription of the disease. The knowledge base is imple-
mented in OWL (Bechhofer et al., 2009). The Extrac-
tion Manager is in charge of the extraction and initial
ICEIS2012-14thInternationalConferenceonEnterpriseInformationSystems
154
preprocessing of medical records from the EHR sys-
tem. This component is parametrized according to the
characteristics of the system and extracts the records
according to the definition of initial parameters, such
as date of admission, gender or age of patients. Per-
sistent Manager and the DataStore Manager store the
required tables to perform the search process inside
a database. These tables are used to create a single
view with all the unstructured and structured data.
The component Text Mining is the core of the anal-
Figure 3: DISEArch component diagram.
ysis and implements Stemming using Porter Stemmer
algorithm, simple string tokenisation, sentence split-
ting, POS tagging using Probabilistic Part-of-Speech
Tagging Using Decision Trees (Schmid, 1994) for an-
notating text with part-of-speech and lemma informa-
tion and finally gazeteer lookup using regular expres-
sions. This component has a coordinator that calls
each of the search engines. The Narrative Search En-
gine is in charge of the analysis of natural language
and was developed using the GATE API (Cunning-
ham et al., 2011). This API enables the inclusion
of all the language processing functionality within
DISEArch. In addition, we use Treetagger (Schmid,
1994), a Pearl implementation which provides tok-
enization and Part of the Speech tagger. The Struc-
tured Search Engine is in charge of searching the dis-
ease over the structured attributes. Finally the Inte-
grator component integrates the results using the se-
mantic rules and prioritizes the set of records.
4.2 Experiment Context and Results
Pulmonary Embolism (EP) was chosen to test DIS-
EArch. A medical expert provided the subgroups
and literals that describe it. Preliminary selection pa-
rameters for EHRs were defined: patients over 18
years old and records created between 2009-2011.
One key item to obtain precision and recall was the
prioritization process that was explained in Section
3. The results and their associated medical records
were clustered according to their relevance (Lowly
prioritized, Mildly prioritized and Highly prioritized
medical records). The obtained results with DIS-
EArch were 250 medical records, which correspond
to records with at least one positive literal w.r.t the
disease description. From these records, the priori-
tization process classified 30 as high, 52 as medium
and 168 as low, according to the distance function.
In order to validate the precision and recall of
DISEArch a medical expert analysed manually the
records detecting 112 EP positive medical records.
From these results DISEArch obtained 30 as high,
50 as medium and 32 as low. The precision and re-
call are presented in Figure 4. As expected the preci-
sion and recall is high for high and medium positive
records. The low precision of the Low group is the
consequence of the inclusion of records that contain
few literals in common with the disease template.
Figure 4: Precision and recall of DISEArch.
5 CONCLUSIONS
The DISEArch strategy presented in this paper en-
ables medical researchers to identify those EHRs that
include the diagnosis of a specific disease. This time-
consuming and expert-dependent task can be sup-
ported by DISEArch through specific rules for iden-
tifying diseases and weights to prioritize the selected
records, leaving the expert task to one of review and
acceptance, rather than search and retrieval. DIS-
EArch goes beyond classical text mining because it
uses unstructured text in medical records as well as
related structured fields to enrich the final results.
From our first tests we found that, although the non-
prioritized results are already helpful and accurate
(as compared to expert selected records), prioritiza-
tion still plays an important role in the classifica-
tion of medical records because it adds precision and
contributes to the review process by presenting the
records in terms of how close they are to the disease
template.
DISEArch-AStrategyforSearchingElectronicMedicalHealthRecords
155

ACKNOWLEDGEMENTS
This work was supported by the project “Iden-
tificaci
´
on semiautom
´
atica de pacientes con enfer-
medades cr
´
onicas a partir de la exploraci
´
on retro-
spectiva de las historias cl
´
ınicas electr
´
onicas reg-
istradas en el sistema SAHI del Hospital San Ignacio”
made by Pontificia Universidad Javeriana and Hospi-
tal Universitario San Ignacio.
REFERENCES
Antal, P., de Moor, B., and M
´
esz
´
aros, T. (2001). Anno-
tated bayesian networks: A tool to integrate textual
and probabilistic medical knowledge. In Proc. of the
14th IEEE Symp. on Computer-Based Medical Sys-
tems, CBMS ’01.
Averbuch, M., Karson, T. H., Ben-Ami, O., and Rokach,
L. (2004). Context-sensitive medical information re-
trieval. Studies in health technology and informatics.
Bechhofer, S., van Harmelen, F., Hendler, J., and Horrocks,
I. (2009). ”owl web ontology language reference”.
Technical report, W3C.
Breault, J. L., Goodall, C. R., and Fos, P. J. (2002). Data
mining a diabetic data warehouse. Artificial Intelli-
gence in Medicine.
Chapman, W. W., Bridewell, W., Hanbury, P., and Cooper
(2001). A simple algorithm for identifying negated
findings and diseases in discharge summaries. J. of
Biomedical Informatics.
Claster, W., Shanmuganathan, S., and Ghotbi, N. (2008).
Text mining of medical records for radiodiagnostic
decision-making. JCP.
Cunningham, H., Maynard, D., Bontcheva, K., Tablan, V.,
and Aswani, N. (2011). Text Processing with GATE
(Version 6).
Ginter, F., Suominen, H., Pyysalo, S., and Salakoski, T.
(2009). Combining hidden markov models and latent
semantic analysis for topic segmentation and labeling:
Method and clinical application. I. J. Medical Infor-
matics.
Han, H., Choi, Y., Choi, Y. M., Zhou, X., and Brooks, A. D.
(2006). A generic framework: From clinical notes to
electronic medical records. Computer-Based Medical
Systems, IEEE Symp.
Hanauer, D. A. (2006). Emerse: The electronic medical
record search engine. AMIA A. Symp Proc.
Hotho, A., N
¨
urnberger, A., and Paass, G. (2005). A brief
survey of text mining. LDV Forum.
Huang, M.-J., Chen, M.-Y., and Lee (2007). Integrating data
mining with case-based reasoning for chronic diseases
prognosis and diagnosis. Expert Syst. Appl.
Keeney, R. and Raiffa, H. (1976). Decisions with multiple
objectives: Preferences and value tradeoffs. J. Wiley,
New York.
Manning, C. D., Raghavan, P., and Schtze, H. (2008). In-
troduction to Information Retrieval.
Rokach, L., Romano, R., and Maimon, O. (2008). Negation
recognition in medical narrative reports. I. R.
Schmid, H. (1994). Probabilistic part-of-speech tagging us-
ing decision trees. In Proceedings of the International
Conference on New Methods in Language Processing.
Seyfried, L., Hanauer, D. A., Nease, D., and Albeiruti
(2009). Enhanced identification of eligibility for de-
pression research using an electronic medical record
search engine. Inter. J. of Medical Informatics.
Spasic, I., Ananiadou, S., McNaught, J., and Kumar, A.
(2005). Text mining and ontologies in biomedicine:
Making sense of raw text. Briefings in Bioinformat-
ics.
USNLM (2011). Unified medical language system
R
(umls
R
). http://www.nlm.nih.gov/research/umls/.
Noviembre 25, 2011.
Zhou, X., Han, H., Chankai, I., Prestrud, A. A., and Brooks,
A. D. (2005). Converting semi-structured clinical
medical records into information and knowledge. In
Proc. of the 21st Inter. C. on Data Eng. WS.
ICEIS2012-14thInternationalConferenceonEnterpriseInformationSystems
156
