
Nonlinear Deterministic Methods
for Computer Aided Diagnosis in Case of Kidney Diseases
Andreea Udrea, Mihai Tanase and Dumitru Popescu
Department of Automatics, University Politehnica of Bucharest, Bucharest, Romania
Keywords: Computer Aided Diagnosis, Nonlinear Deterministic Methods, CT Images.
Abstract: This paper proposes a set of nonlinear deterministic methods derived from chaos theory that can serve as
computed aided diagnosis tools for kidney diseases based on computer topographies (CT). These procedures
target the classification of the analyzed tissue samples in normal, malign and benign affected and also
enhanced visualization of the CT images. The classification methods consist in estimating the fractal
dimension of the kidney tissue and, respectively, the correlation dimension of the attractor obtained from the
spatial series associated to the kidney image. The enhanced visualization method associates a fractal map to
the analysed image. The methods are tested on 120 CTs presenting normal and modified tissue. The degree
of trustworthiness of the methods while dealing with classifications is discussed based on statistical results
and samples of fractal maps associated to the images are also presented.
1 INTRODUCTION
In order to increase the life expectancy and improve
the overall quality of life for patients with kidney
diseases, a critical stage in the medical process is to
employ a suitable protocol for delivering the
diagnostic, establish a treatment and, when needed,
to design an appropriate follow up procedure.
Usually, the first investigations and the follow up
consist in noninvasive or minimally invasive
procedures, in order to obtain the biological data for
proposing an accurate diagnostic. In this context,
any improvement in interpreting the patient’s data is
highly important.
When considering such data, at least two main
problems of tremendous importance have to be
solved: capturing and storing the considered medical
signals, on one hand and, on the other hand,
analyzing and interpreting the stored signal. In order
to capture 2D signals of the kidneys the most used
procedure is computer tomography (CT). The
obtained signals (grey level images) are usually
highly nonlinear, rather noisy and due to close
values of radio densities of the tissues, sometimes
difficult to interpret.
Analysis and interpretation of the captured
medical signals is almost exclusively subjected to
the human diagnosis expertise and experience. In the
last decade, a lot of effort was made to create
automatic analysis and diagnosis tools for aiding the
medical act. Nowadays, automatic diagnostic is still
a long term goal to be achieved, but Computer-
Aided Diagnosis (CAD) systems design seems
possible. This is confirmed by all major medical
imaging companies increasing interest in developing
CAD systems. Three signal processing operations
are closely related to CAD topics: filtering,
segmentation and quantification of analyzed
features. Enhanced visualization is another
important aspect, especially in the context of kidney
CT images presenting benign affected tissue.
The main goal of this paper is to present a series
of new or improved nonlinear methods that can aid
the medical diagnostic in the case of kidney diseases
and can be included in a CAD system. These
methods are derived from nonlinear time series
analysis and fractal geometry, both branches of
chaos theory. Their primary goal is to analyze the
kidney CT images and to decide if the presented
tissue is normal, malign affected, benign affected
and also to try differentiating between types of
benign diseases and malignancy stages. Where these
classification attempts prove limitations, enhanced
visualization procedures are offered as support.
The paper is organized in four chapters. In what
follows the nonlinear deterministic methods to be
used are presented and the results obtained by
employing them to the analysis of CTs are
511
Udrea A., Tanase M. and Popescu D..
Nonlinear Deterministic Methods for Computer Aided Diagnosis in Case of Kidney Diseases.
DOI: 10.5220/0004039405110516
In Proceedings of the 9th International Conference on Informatics in Control, Automation and Robotics (ICINCO-2012), pages 511-516
ISBN: 978-989-8565-21-1
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

statistically analyzed. For this study, a series of 120
CT images were used: fifty of them contain malign
modified kidney tissue, fifty images present normal
kidney tissue and the rest of twenty are organised in
four images groups of benign affections. In the end
conclusions are summarized.
2 THE PROPOSED NONLINEAR
DETERMINISTIC METHODS
FOR CAD
Nonlinear time series analysis (NTSA) and fractal
analysis, as branches of chaos theory, provide useful
methods for the characterization of mono and multi
variable signals (like time series and images).
2.1 Attractor’s Correlation Dimension
Estimation Method for the CT
Image Associated Time Series
Typically, the NTSA deals with series that are sets
of values of a single variable function, usually
measured as function of time (dynamic features).
Nonlinear methods have been developed in the past
20 years, being motivated by the concept of
deterministic chaos, which is proved to exist within
many real systems in biology, medicine, chemistry,
physics and electronics. The studied time series in
medicine and biology are: recordings of the
electrical activity – electrocardiograms (EKG),
electroencephalograms (EEG) and physiological
parameters – blood pressure, pulse, and breathing
rate. Here are some applications with important
results of NTSA: diagnosis and control of cardiac
arrhythmia and ventricular fibrillations prediction
(Perc, 2005); characterization of sleep stages
(Rajendra, 2005); evaluation of variations in brain
functioning for psychical processes characterization
(Mekler,2008; Pritchard and Duke 1995);
characterization of anesthesia state (Widman, 2000).
The proposed NTSA derived method is based on
the invariant measure of a chaotic dynamical system:
the correlation dimension of the system’s attractor.
By investigating time series, one can observe the
behavior and properties of dynamical systems, in our
case of different physiological parameters.
From the mathematical point of view, there is a
formalism to describe the time series features. Let
the real valued map F :M →R be a measure on the
state space of some discrete dynamical system T
providing data in M. If s>0 is a fixed delay
(assigned to some sampling period) and x is a fixed
state, then a time series is a sequence of
measurements like the following:
F (T(t,x)), F (T(t + s,x)), F (T(t + 2s,x)),
…, F (T(t + (N −1)s,x))
(1)
for any starting instant t . Note that the state changes
too during the time series acquisition. The samples
of time series are often simply denoted by x
t
, x
t+1
,
x
t+2
, … .In context of medical signals, these time
series are measurement (like EEG, EKG) performed
on some patient (represented here by the system T ).
The acquisition rate and the length of the
measurement depend on the type of investigated
parameter. One can reconstruct the attractor of a
dynamical system from the time series generated by
the system, by using the Taken’s Embedding
Theorem (Taken, 1981) and computing the
correlation dimension of the attractor in order to
geometrically characterize it. The correlation
dimension d
C
is calculated using the following
recipe:
0
ln ( )
() , 0 lim
ln
C
d
C
C
Cd
ε
ε
εεε
ε
→
=→⇒=
(2)
where C(ε ) is the correlation integral defined below:
2
,1
1
() lim ( | |)
N
ij
N
ij
CHyy
N
εε
→∞
=
=−−
∑
(3)
and: H is the Heaviside step function (which returns
either the unit value for non negative arguments or
null value otherwise), ε is the accepted distance
between points, y
i
is a point in the embedded phase
space constructed from a single time series,
according to Taken’s theorem, i.e:
y
i
=(x
i
,x
i+s
,x
i+2s
,…x
i+(dE-1)
s), s is the delay, d
E
is the
dimension of the embedding space where the
attractor resides, N is the number of embedding
vectors. So, C(ε ) gives the proportion of number of
points couples in the embedding space with the
Euclidian distance less than a specified small
threshold ε .
In pathology (especially in case of CT, RM
images and frozen tissues samples), one deals with
static (invariant) structures. In this case,
measurements are taken with respect to the one-
dimensional spatial axis, instead of temporal axis. In
this context we propose a method for reconstructing
the attractor from a CT image an associate to it a
specific d
C
.
In order to perform nonlinear analysis on a CT
normal or modified tissue image, a series of steps
must be made.
ICINCO2012-9thInternationalConferenceonInformaticsinControl,AutomationandRobotics
512

First, from a CT slice, the region containing the
tissue to be analyzed must be isolated; a matrix
containing values of each pixels shade is obtained
(the value can vary between 0 and 255
corresponding to different shades of grey; 0 stands
for black and 255 for white). The time (spatial)
series is generated in the following manner: the
matrix resulting from the original image is cut in
horizontal strips of 1, 2, 4, 8, … pixels, with respect
to the initial image dimension and precision; all
strips are put together one after another and generate
one single strip associated to the image; the time
(spatial) series - x(t) - is generated by computing
either the mean value or the maximal (dominant)
value of each column of pixels within the strip.
As result of this procedure, the time (spatial)
series associated to the section of the analyzed tissue
is obtained. For this study, since the analyzed CT
regions are not extremely large, a 1-pixel strip was
associated to each original image, this way not
altering the information provided by the image.
Having the associated series, the next step of the
procedure implies calculating the correlation
dimension of the attractor. This value is the
discrimination criterion.
However, in practical applications, in order to
determine the dimension of an attractor, we cannot
directly use the above formulae for d
C
due to the
following aspects: limited time series; noisy time
series; unknown fractal dimension of the attractor;
for different s - delay values different results due
autocorrelations; unknown d
E
– leading to time
correlations when reconstructing the series in a
embedding space with unsuitable dimension; time
series with the first part of data not on the attractor.
The delay or lag value -s- used to create the
delayed embedding must be properly chosen (Kantz
and Schreiber, 2003). A small value of the delay
generates correlated vector elements, while large
delay values yield to uncorrelated data and a random
distribution in the embedding space. The delay can
be chosen with good results as the moment of time
where the autocorrelation function of the
reconstructed series decays to 1/e of its initial value:
() (1)(1 1/)
R
NRN e
τ
<−
.
(4)
Generally, the lag value is found between 4 and
10, while the used search interval is [1, 20].
The minimum allowed embedding dimension is
the dimension where the number of so called false
nearest neighbours drops under a certain percent. A
false neighbour is a point that under a certain higher
dimensional embedding is projected near a point that
in the previous embedding is not in its vicinity.
In order to implement this procedure, each point
of the delayed series is tested by taking its closest
neighbour in d
E
dimensions, and computing the ratio
of the distances between these two points in d
E
+1
dimensions and in d
E
dimensions. If this ratio is
larger than a certain threshold th, the neighbour is
false (this threshold is taken large enough to take in
consideration points that exponential diverge due to
deterministic chaos):
11
,,
,,
EE
EE
id jd
id jd
yy
th
yy
++
−
>
−
(5)
where ||.|| is the Euclidian distance.
The percentage of false neighbours is computed
over a range of embedding dimensions (d
E
between 2
and 15) until it reaches a value less than a specified
limit; otherwise it considers the minimal obtained
value.
Once a proper delay and a minimum allowed
embedding dimension are determined, the
correlation dimension is calculated over a range of
different
ε
- values and embedding dimensions
higher than the first assuring a decreased number of
false neighbours.
The d
C
differs from one embedding dimension to
another due to the noise in the data, but there is a
particular region, usually called the scaling region
where d
C
stabilizes (Kantz and Schreiber, 2003).
This is the interval where a mean value for the
correlation dimension of an attractor is calculated.
2.2 The Box-Counting Dimension
Estimation Method
Fractal analysis methods are used for the description
and quantization of geometric features of irregular
forms and patterns. Its most known tool is the fractal
dimension used to provide information on the
irregularity of an object contour or self-similarities
of a texture, which associates to some pathology as
well. It was applied for the study of medical systems
and subsystems at microscopic and macroscopic
scale, fracture analysis or texture classification
(Peitgen, 1992). The simplest medical application
consists in the morphological analysis of a structure
(for example, the lung network of arteries and
veins). This analysis of irregularities can be applied
in a similar manner on different forms, like the
delimitation between normal and affected tissue,
lesions, and tumors.
Here are some examples of fractal analysis
results in medicine and biology: classification in
pathology (Bassingthwaighte, 1994; Dobrescu si
NonlinearDeterministicMethodsforComputerAidedDiagnosisinCaseofKidneyDiseases
513

Vasilescu, 2004) and physiology (Luzi, 1999), tumor
growth description (Landini, 1998). The box
counting method provides a measure of fractal
dimension d
f
estimated for texture or contour.
The d
f
, derived from the Hausdorff coverage
dimension, is given by the following approximation:
()
()
0
log ( )
lim
log 1/
f
s
Ns
d
s
→
=
(6)
where: - N(s) is the number of squares with side
length s that contain information when grid covering
the image.
Relation (6) is the equation of the slope d
f
, of the
regression line associated to the points (log(N(s),
log(1/s)) for different values of the square’s side – s
of the covering grid..
The standard Box-Counting algorithm assumes
to determine the d
f
in accordance with the
dependence of the texture upon the used scale factor.
It consists transforming the grey scale image in
binary image, successively covering it with a grid of
squares of equal sides (2, 2
2
, 2
3
, ..) and counting each
time the squares that contain some part of the
analyzed object. The points of coordinates
(log(N(s)), log(1/s)) are approximately positioned in
a line and its slope is the fractal dimension in “box-
counting” sense.
To exemplify how the algorithm is used, we’ll
consider the image of a kidney (Figure 1. a)) from
which we’ll extract a binary version by neglecting all
the pixels over a certain threshold (Figure 1. b)).
a) b) c)
Figure 1: a) The original image; b) binary image
c) extracted contour.
Next, we’ll apply the box-counting algorithm,
described above, for different scale values s.
This method can be also used to determine the
self similarities of an object contour (Figure1 c)), but
in our case, due to the fact that the kidney capsule is
not necessary affected, the texture is more important.
A general problem of this method is the use of an
ad hoc threshold when creating the binary image.
This fact leads to incomplete or “noisy” object in the
binary image and sometimes importantly affects the
fractal dimension value.
2.3 Weighted Box-Counting Dimension
for Image Enhancement
This algorithm is based on the fact that in the CT
images a higher density of the tissue is equivalent to
lighter gray. Our idea was to associate to every pixel
a weight proportional to its gray level. We resume
the essential of the algorithm below.
Let us consider an image. We cover the image
with square boxes as in the standard Box-Counting
algorithm. Let
k
s be the size of the box used in
covering at step
k
(therefore we have to compute
)(
k
sN at this step). Let ),( yx be the coordinate
of the upper-left corner of one of these boxes (let
this be the box
k
t
B ).
We now define
k
t
m as the maximum of the
weight values of the pixels contained in this box.
(7)
where
ji
w
,
is the weight associated to the pixel at
(i,j) coordinates.
Let
k
tk
k
t
k
t
rsmW += ]/[
, where if
k
tk
ms |
then
1=
k
t
r else 0=
k
t
r .
Therefore
∑
=
t
k
tk
WsN )( .
Next, the computation formula for d
w
is the
similar to the one in the classical algorithm. We
shall refer to the number d
w
as the Weighted Box
Counting Dimension or WBCD. Let us consider an
image and let A be a pixel on it. Let K be a square
centered at A. By using the previous algorithm we
compute the WBCD of the square K and we
associate a color to the pixel A according to this
WBCD (the function which associates the color is a
key part of the algorithm). This way we obtain a
map of level lines (we shall refer to this map as the
Fractal Map or FM).
This leads to a classification of different tissues
according to the associated color. Different structures
must have different colors. The use of the FM in
diagnosis requires a database with sufficient images.
3 RESULTS AND STATISTICS
We start the analysis procedure by presenting the
statistical results obtained by using d
F
and d
C
as
ICINCO2012-9thInternationalConferenceonInformaticsinControl,AutomationandRobotics
514
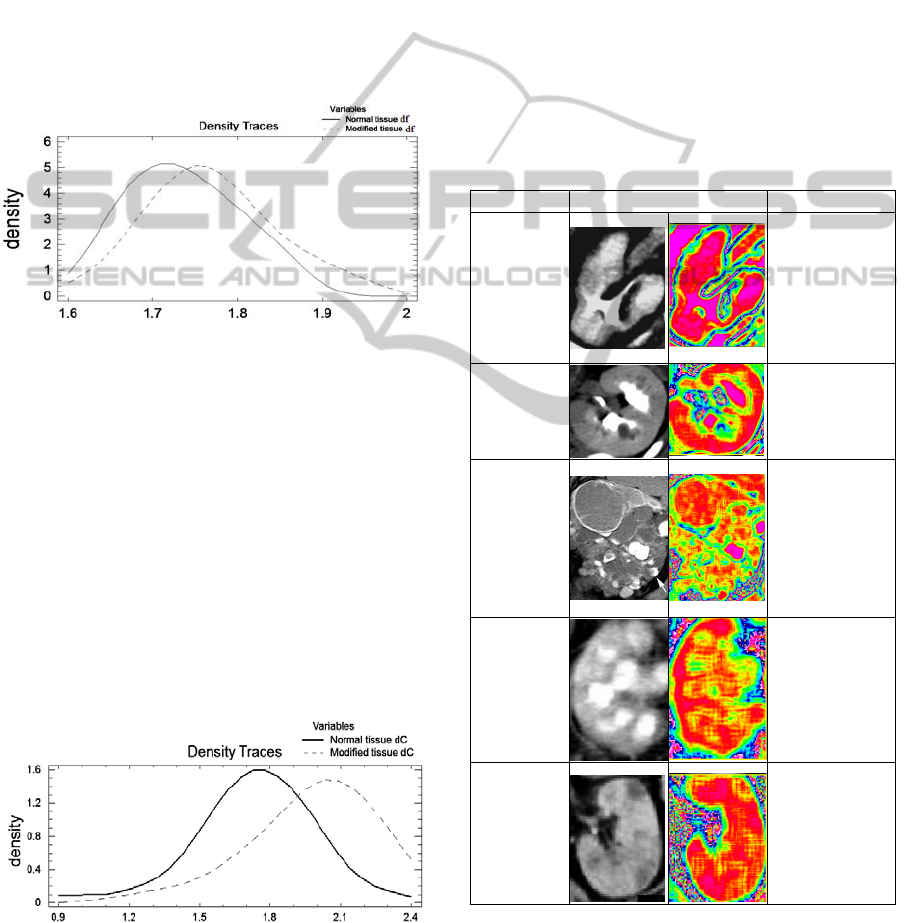
classification methods. One hundred and twenty CT
images were analysed; they were divided into two
equal samples: containing normal tissue and half
modified tissue.
For the statistical analysis, descriptive and
comparison procedures were performed. For each
sample, the average, standard deviation, standard
skewness and standard kurtosis were computed.
In order to compare the samples the t test and
Kolmogorov-Smirnov test were performed.
In the case of d
f
both comparison tests show no
significant difference between the two distributions
at the 95.0% confidence level. So, the
trustworthiness of this classification is low.
Figure 2: Comparison of density traces.
In the case of d
C
both comparison tests show
significant difference between the two distributions
at the 95.0% confidence level.
The average is 1.729 for the normal tissue and
respectively 1.974 for the modified tissue.
The 95.0% confidence interval for mean is
[1.6282,1.83155] for the normal tissue and
[1.87225,2.07725] for the modified tissue.
We conclude that the box-counting method is
using a certain threshold, this way loosing some
information on the tissue texture while nonlinear
analysis is more precise and uses all the information
in the images. We recommend the use of the second
method for analyzing CT images.
Figure 3: Comparison of density traces.
We have also compared the results acquired
when the CT was taken with contrast substances and
without. In the second case, the d
C
values are smaller
because of a series of features that are not so visible
(blood vessels). The differences between the d
C
of
normal and modified tissue samples are smaller. So,
we suggest that this methodology is better to be used
with associated time series resulting from CT images
taken with contrast substances.
The second step in the analysis was to determine
the correlation dimension of the attractor for images
containing kidneys with benign affections.
The discrimination is obvious in the cases of
pyelonephritis (the resulted d
C
values being smaller
than in the case of normal tissue) and kidney
tuberculosis (with
d
C
values larger than in the case
of malign modified tissue).
Table 1: Benign modified tissue images, their fractal maps
and associated d
C
values.
Affection Image Enhanced d
C
Pyelone-
phritis
1.36(correspo
ndent d
C
for
healthy kidney
-1.85)
Medullary
sponge
kidney
1.91(1.86)
Polycystic
kidney
2.06 (1.9)
Kidney
tuberculos
is (renal
TB)
2.3(1.92)
Thrombosi
s
1.98(1.93)
The d
C
values for medullary sponge kidney
tissue and thrombosis affected kidney tissue are
generally a little bit larger than the ones for normal
tissue.
NonlinearDeterministicMethodsforComputerAidedDiagnosisinCaseofKidneyDiseases
515

The d
C
values for polycystic kidney tissue were
generally larger than the ones for normal tissue.
In the third column of the above table the kidney
CT image fractal map is presented.
The kidney border and affection specific aspects
like different types of tissue clusters and their
delimitation can be seen clearer.
Also, the different colours in the map identify
different formations, specific to the affection.
This method proved more useful than the
previous two in aiding the diagnostic in the case of
benign affected tissue.
We conclude that discrimination between these
benign affections can be done but needs a larger
database of images.
Further work will focus on enlarging the CT
images data base in order to provide more accurate
discrimination interval values for different types of
kidney affections.
4 CONCLUSIONS
The conclusions of the study on the selected set of
CT images are: there are significant differences
between the correlation dimension of the normal
tissue and the correlation dimension of the modified
tissue; significantly better results are obtained in the
case of CT images taken when contrast substances
are used. The proposed nonlinear method for
estimated the correlation dimension associated to a
CT image proved efficient for differentiating
between normal and modified kidney tissue while
the box-counting method failed in providing useful
results. The image enhancement method proved very
helpful when inconclusive classification was
obtained for benign tissue.
Future work aims at: enlarging the CT images
data base; creating the fractal model of the kidney,
measuring, where it is possible, the percentage of the
modified tissue in a kidney CT slice in order to
provide information on what is causing the increase
in d
C
(percentage of affected tissue or d
C
value of
modified tissue); determining the position of masses
in an affected organ when considering horizontal
slices and respectively reconstructed transversal
slices in that organ .
REFERENCES
Bassingthwaighte J. B., Liebovitch L. S., West B. J.,
1994. Fractal Physiology. Oxford University Press
Dobrescu R., Vasilescu C., 2004. Interdisciplinary
applications of fractal and chaos theory. Romanian
Academy Press, pp. 247-254, 2004
Luzi P., Bianciardi G., Miracco C., De Santi M.M., Del
Vecchio M., Alia L., Tosi P., 1999. Fractal analysis in
human pathology. Ann. N.Y. Acad. Sci. 879, 255-257
Landini G., 1998. Complexity in tumor growth patterns,
Fractals in Biology and Medicine. Birkhäuser Verlag,
pp. 268-284
Kantz H., Schreiber T., 2003. Nonlinear time series
analysis, 2003. Cambridge University Press, 3
rd
edition
Mekler A., 2008. Calculation of EEG correlation
dimension: Large massifs of experimental data.
Computer Methods and Programs in Biomedicine,
vol. 92, pp. 154-160
Peitgen H. O., Jurgens H., Saupe D., 1992. Chaos and
Fractals – New Frontiers of Science. Springer-Verlag
Perc M., 2005. Nonlinear time series analysis of human
electrocardiogram. In European Journal of Physics,
vol. 26, pp. 757-768.
Pritchard W. S., Duke D. W., 1995. Measuring chaos in
the brain: A tutorial review of EEG dimension
estimation. Brain Cognition, vol. 27, pp. 353-397
Rajendra U., Faust O., Kannathal N., Chua T.,
Laxminarayan S., 2005. Non-linear Analysis of EEG
Signals at Various sleep stages. In Computer Methods
and Programs in Biomedicine, vol. 80, pp. 37-45.
Takens F., 1981. Detecting strange attractors in fluid
turbulence. In Dynamical System andTurbulence,
Rand D. A., Young, L. S. (Eds), Lecture Notes in
Mathematics, Springer Verlag
Widman G., Schreiber T., Rehberg B., Hoerof A., Elger C.
E., 2000. Quantification of depth of anesthesia by
nonlinear time series analysis of brain electrical
activity. Physical Review E, no. 62, pp. 4898-4903
ICINCO2012-9thInternationalConferenceonInformaticsinControl,AutomationandRobotics
516
