
An Ontology-based Data Acquisition Infrastructure
Using Ontologies to Create Domain-independent Software Systems
Dominic Girardi
1
, Klaus Arthofer
2
and Michael Giretzlehner
1
1
Research Unit Medical-Informatics, RISC Software GmbH, Hagenberg, Austria
2
Department for Process Management in Health Care, Upper Austria University of Applied Sciences, Steyr, Austria
Keywords:
Ontology, Meta Modelling, Data Acquisition.
Abstract:
We created an ontology-based data acquisition infrastructure which is able to store data of almost arbitrary
structure and can be set up for a certain domain of application within hours. An ontology editor helps the
domain expert to define and maintain the domain specific ontology. Based on the user-defined ontology, a
web-based data acquisition system and an ETL data import interface are automatically created at runtime.
Furthermore, rules for semantic data plausibility can be established in the ontology to provide semantic data
quality for subsequent processing of the collected data. After a comprehensive requirement analysis we de-
cided to use a special meta model instead of standard OWL ontologies. In this paper, we describe our meta-
model and the reason for not using OWL in our case in detail as well as we present the infrastructure and the
project it is currently used for.
1 INTRODUCTION
Research in common and medical research in partic-
ular is grounded on a vast amount of data. This pre-
cious data, which is often yielded by expensive stud-
ies or experiments, needs to be stored in a structured
and save way. Since data structures are often com-
plex, a professional data acquisition system is needed
to acquire and store the data. Due to the fact that
every field of study requires its own individual data
structure, these data acquisition systems are hardly
reusable and need to be developed individually for
each new domain, which is elaborate and expensive.
For that reason study authors tend to use sub-optimal
data storage solutions like excel sheets.
In order to encounter this drawback, we developed
an ontology-based data acquisition and storage infras-
tructure including a generic web-based data acquisi-
tion system, an ontology-guided ETL module, an easy
to use ontology editor, and a powerful rule checking
mechanism to ensure semantic data quality. It is able
to store data of almost arbitrary structure and ready to
be used for a certain domain within a few hours.
In Section 3 we describe the technical background
of the system in detail including the ontology format
we use. In Section 4 we motivate, why we did not use
the standard ontology format OWL for our system and
oppose OWL to our meta model. Section 5 contains
the practical results of the projects are presented in-
cluding screen shots and key numbers of the running
system. In Section 6 we summarize our results.
2 RELATED RESEARCH
The most related work to ours is described in (Zavaliy
and Nikolski, 2010). In their paper - that only con-
tains 1 page - the authors describe the use of an on-
tology for data acquisition, motivated by the demand
of adaptive data structures. They used an OWL on-
tology to model their domain, which consists of four
concepts (Person, Hospital, Diagnosis and Medica-
tion). There is no information given on user inter-
faces or semantic data checks. Asides from this work,
it was very hard to find any publications on ontology
based data acquisition systems. In most publications
ontologies are used for information extraction from
text (Tran and Kameyama, 2007) or to enrich the ex-
isting data with structural and semantic information
or to build a cross-institutional knowledge base. In
(Kataria et al., 2008) the authors describe the usage
of ontologies for inter-hospital data extraction to im-
prove patient care and safety by analyzing hospital ac-
tivities, procedures, and policies. Here, the ontology
is strongly connected to the project. In (Kiong et al.,
2011) e.g. the authors describe an ontology based sys-
155
Girardi D., Arthofer K. and Giretzlehner M..
An Ontology-based Data Acquisition Infrastructure - Using Ontologies to Create Domain-independent Software Systems.
DOI: 10.5220/0004108101550160
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2012), pages 155-160
ISBN: 978-989-8565-30-3
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)
tem for extracting research relevant data out of het-
erogeneous hospital information systems. Here again,
the purpose is to find a superior common data model
to compare data of different medical institutions. In
contrast to our system, the ontology is strongly con-
nected to the project.
3 META MODEL-BASED DATA
ACQUISITION SYSTEMS
State-of-the-art data storage and acquisition systems
are logically organized in multiple layers (Balzert,
2010). This multi-tier architecture - mostly com-
mon is a three tier architecture with a data layer, a
logical layer and a user interface layer - helps to re-
duce the technical dependencies among the particular
layers and makes them exchangeable as long as the
interfaces between the layers remain constant. Still,
the semantic dependency of the whole system from
the data structure of the field of application - the real
world problem - cannot be resolved by using multi-
tier-architectures. Changes to the data model, caused
by changing requirements, cause transitive changes to
the whole application. This makes the whole system
rigid, inflexible and only adaptable at high costs and
personal effort. In order to resolve this dependency
a special data model is needed. It must be indepen-
dent from the field of application, which sounds self-
defeating at a first glance. How should a data model
store data for a real world domain, without being in-
fluenced by it? Meta models are data models of higher
level of abstraction and can be used to solve this issue.
3.1 Meta Models
The Object Management Group OMG (MOF, 1997)
defines four levels (M
0
- M
3
) of meta modeling. Each
model at level M
i
is an instance of a model at level
M
i+1
. Level M
0
contains the real world transactional
data. Each object at M
0
is an instance of a model
defined in M
1
, which is called the model layer. Each
model at level M
1
can be seen as an instance of a meta
model at level M
2
- the meta model layer. Level M
3
contains meta meta models.
Conventional data storage systems use M
1
data
models, which directly describe the field of applica-
tion. M
2
meta models describe M
1
data models. We
developed a meta model which is able to store M
1
data
models as well as their M
0
transactional data. This al-
lows replacing the conventional M
1
data model by the
M
2
meta model. As a consequence the logical layer
cannot be implemented directly anymore, but serves
as an execution enviroment for the logic rules, which
are stored in the meta model. Furthermore, the user
interface(s) can be created automatically based on the
meta information stored in the meta model.
3.2 Meta ER Model
Since most M
1
data models are relational models
stored in a relations database, they can be described
using an Entity Relationship model (ER model),
which was introduced by Peter Chen in 1976 (Chen,
1976). Consequently a meta model, which is able
to describe an ER model can be used to store data
structures that are stored in a relational database. Our
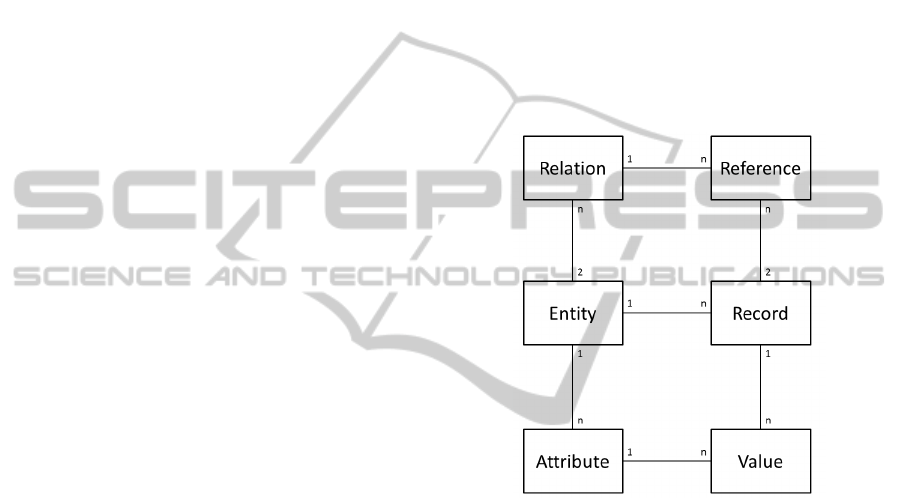
meta ER model consists of six main entities which are
shown in Figure 1.
Figure 1: ER Diagram of our Meta ER data model.
The central entity class is Entity. It corresponds
to a table in a relational database. So, for each table
that is needed in the M
1
data model a row in the En-
tity table is created. Thus, the independence of the
data model from the real world is achieved. Chang-
ing entities do not cause changes to the data model
any more, but only to its content. The same mecha-
nism applies to attributes and relations. If an attribute
is needed, it is simply added to the Attribute table in-
stead of creating a new attribute to a table and chang-
ing the data model itself. While the entity classes
on the left hand side describe structural elements, the
transactional data is stored in the three entity classes
on the right hand side: Record, Value, and Reference.
A table in a relational database consists of columns
(attributes) and rows. Each row is a single record of
a certain entity. Each single cell in a table contains a
value which can be uniquely assigned to exactly one
record and one attribute of an entity. In addition to
these basic entities the whole meta data-model con-
sist of 19 more entity classes, which are responsible
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
156

for, auditing, user management, GUI customization,
and semantic plausibility checks.
4 WHY WE DID NOT USE OWL
ONTOLOGIES
So far, our basic meta model does not provide any
feature that cannot be covered by classical OWL on-
tologies. According to (Chandrasekaran et al., 1999)
”Ontologies are content theories about the
sorts of objects, properties of objects, and re-
lations between objects that are possible in a
specified domain of knowledge.”
(Gruber, 1993) provides a more general definition:
”An ontology is a specification of a conceptu-
alization.”
Their original purpose is to share a common knowl-
edge of a domain across multiple institutions. An as-
pect which is meant to be used as a backbone for the
semantic web (Ding, 2001) and (Berners-Lee et al.,
2001). Furthermore, ontologies are very popular for
knowledge representation and organization in many
scientific domains like the Gene Ontology (Smith
et al., 2003) in bioinformatics. According to the defi-
nitions above, meta models can be seen as equivalent
to ontologies. Consequently the question arises, why
we did use our own meta model instead of the stan-
dard ontology format OWL, since there exist numer-
ous ontology editors like Protege project (Knublauch
et al., 2004) and reasoning engines.
4.1 OWL
The Web Ontology Language OWL is a XML based
description language for ontologies (McGuinness and
van Harmelen, 2004). It is based up the Resource
Description Framework (RDF) (Manola and Mille,
2004) and part of the Semantic Web Stack. The main
concepts of OWL are classes, properties and individ-
uals (Horridge et al., 2004).
Individuals represent objects in the real world
domain this ontology models. They correspond to
records in our meta model, with the difference that
OWL individuals can be part of more than one OWL
class, which is not possible in our meta model, where
each record is an instance of exactly one entity class.
OWL properties are binary relations which connect
two individuals (or values), which corresponds to re-
lations in the ER model or to a data type which can be
expressed using attributes in the ER model. Classes,
comparable to entity classes in the ER world and in
our meta model, are interpreted as sets containing in-
dividuals. They encapsulate all properties that can be
applied to these individuals. Basically, all concepts
in the ontology can be mapped to a corresponding
concept in our meta model, whereas their behavior
is sometimes different. For our purpose the tradition
restricted behavior of the ER world is the more suffi-
cient one. The degree of freedom in OWL is gener-
ally higher than in our meta model. Still, this degree
of freedom doesn’t always make sense for a data ac-
quisition system e.g. individuals should only belong
to one well defined entity; otherwise they would be
mixed up during subsequent data processing.
4.2 Purpose
OWL Ontologies were not designed to build the back-
bone of generic data acquisition systems. In our sys-
tem the ontology is not just used for data definition
but also as a base for automatic GUI generation. The
information that can be stored in an OWL ontology
would only be sufficient for a very simple GUI design.
For a more customizable GUI more data fields like the
text color, background color, formats, and many more
are needed for each attribute, which are not available
in OWL.
4.3 Reasoning vs. Semantic Plausibility
For OWL ontologies, a variety of reasoners are avail-
able such as Pellet (Sirin et al., 2007), Racer, or
FaCT++. They basically perform inference and rule
consistency checking on the ontology itself. They
check if there is a contradiction within the ontology,
produced by restrictions that are set to the classes of
the ontology. Furthermore, they perform the assign-
ment of individuals to classes. Functionalities which
are indisputably useful, but for our system we needed
something else.
The semantic plausibility rules in our system can
show a structure like this: if a patient is female
and there is at least one treatment for this patient
where the procedure was either an appendectomy (re-
moval of the appendix) or a cholecystectomy (re-
moval of the gallbladder) than the complication of
the type violation of the uterus is plausible, other-
wise it is not. Restrictions like this can hardly be
formulated with OWL syntax. The premise of these
kinds of restriction can only be modeled with defined
classes, whereas for each possible configuration of the
premise-attribute a single defined class has to be mod-
eled - a tasks that seems feasible for the attribute
gender with only two values: male and female, but
hardly doable for an attribute like diagnose with pos-
AnOntology-basedDataAcquisitionInfrastructure-UsingOntologiestoCreateDomain-independentSoftwareSystems
157

sible several thousands of diagnoses. Furthermore, in
OWL, restrictions can only be made to classes and
their properties. Restrictions to transitively connected
classes are not possible. Given a data structure like
this: A patient can contain several treatments and for
each treatment there exist several complications, so
an 1:n relation from patient to treatment and an 1:n
relation from treatment to complication. In OWL it is
very complicated to restrict the possible values in an
individual of the class complication due to the config-
uration of an individual of the class patient. In our
system any attribute of any class can be set into a
dependency relationship to any other attribute of any
other class even if there is no direct connection be-
tween the two classes. This requirement is hardy re-
alizable using OWL syntax and was the main reason
why we decided to use our own meta model instead
of OWL.
4.4 Open World vs. Closed World
OWL ontologies are built upon the Open World As-
sumption. In contrary to the Closed World Assump-
tion, where everything is false, that is not explicitly
defined, the Open World Assumption allows a big de-
gree of freedom in data definition. Although there
are surely good arguments for open world assumption
based software (Baresi et al., 2006) for our applica-
tion the close world assumption is definitely more re-
alistic.
4.5 Storage
While ontologies are usually stored in OWL files, our
system is a database based solution. As mentioned in
Section 3.2 on page 2, aside from the core meta model
entities there exist 19 more entities. Hence, storing
the 19 additional tables in a relation database, while
keeping the ontology information in a file, is a very
inefficient storing strategy. Of course, it is possible to
store an OWL ontology in a relational database (As-
trova et al., 2007), but creating a relation model for
an OWL based ontology is a mix of two concepts that
unnecessarily complicates the system.
5 THE OBIK INFRASTRUCTURE
In 2010 the OBIK project was started with the goal to
create an ontology-based data acquisition system that
is able to store data of almost arbitrary structure. Fur-
thermore, the system should provide a web interface,
which is automatically created based on the ontology
information which allows viewing, entering and edit-
ing the data. Aside from manual data input, electron-
ically stored data should be able to be imported from
heterogeneous data sources by an ontology guided
ETL process. The abbreviation OBIK stands for On-
tology based Benchmarking Infrastructure for Hospi-
tals (German: Krankenanstalten). Although the title
implies a restriction to the field of medicine there is
no such restriction. Since the user is free to design
ontologies for any field of application, this infrastruc-
ture can also be used to collect data for e.g. economic
research or business applications.
The system consists of four main modules OBIK
Management Tool, Web Interface, ETL Plug-In, and
Plausibility Engine.
5.1 OBIK Management Tool
The OBIK Management Tool is a Java-based software.
The main feature is the Ontology Editor (see Fig-
ure 2), which allows the administrator to create and
maintain the ontology. There are no predefined enti-
ties or relations in an empty ontology. So, the admin-
istrator is absolutely free to model the ontology after
the demands of his domain of application. Usabil-
ity had priority to the development of the system, be-
cause the users are experts on their domain, which is
not necessarily ontology design or computer science.
Most operations - such as creation and manipulation
of entities, relations and attributes or the administra-
tion of the plausibility rules - can be done by drag
and drop or by the use of intuitive wizard dialogs.
Furthermore, the administrator can view, manipulate
Figure 2: OBIK Management Tool - Ontology Editor.
and download the stored data. Since the structure of
the presented data depends on the stored ontology, the
structure of the user interface (tables, editors, search
forms) is created dynamically at runtime, based on
the ontology definitions. Changes to the ontology,
have immediate effects on these elements. Numerous
search and filter functions help to extract the desired
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
158

data sets for further processing. The check of plausi-
bility rules can be run from this perspective. Selected
datasets can be downloaded in multiple formats in-
cluding CSV, PDF and XML for subsequent data pro-
cessing. A complete download of the whole ontology
including all data into a relational M
1
data model, ex-
ported as DDL and SQL scripts, is also available.
5.2 Web Interface
The web interface allows the users to enter and edit
data. The interface is primarily used to manually
complement electronically imported data with addi-
tion information. If no electronic sources are avail-
able the whole data need to be entered via the web
interface. Due to the web technology data from dis-
tributed institutions can be brought together in one
central ontology.
The whole structure of the web interface (input
fields, tables, search forms) is derived from the ontol-
ogy and created at runtime. So, changes to the ontol-
ogy have immediate effects to the web forms. Green
and red colored message boxes indicate the result of
the plausibility check for a given data set.
5.3 ETL Plug-in
For importing electronically stored data into the on-
tology the Pentaho Kettle Extraction, Transform,
Load (ETL) tool is used. The powerful ETL tool al-
lows the integration of numerous input sources and
the definition of complex ETL processes. For the last
step, the import of the extracted and rehashed data ta-
bles into the ontology respectively meta model, we
developed a Kettle plug-in. It maps the data fields of
the Kettle data tables onto the structures of our meta
model according to the user’s definitions.
5.4 Plausibility Engine
The Plausibility Engine is responsible for checking
the semantic data integrity. It applies the dependency
rules to each data set and evaluates if the current data
violates any of these. As stated above, dependencies
work on attribute level and set two attributes (master
and slave) in a dependency relation, whereas the cur-
rent value of the slave attribute depends on the cur-
rent value(s) of the master attribute(s). If one slave
depends on more than one master attribute the two de-
pendencies have to be connected using logical opera-
tors AND or OR. Thus, a tree of dependency rules can
be set up for each slave attribute. A visualization of
such a tree can be seen in Figure 3. For the evaluation
process the dependency tree is processed bottom-up
Figure 3: Dependency structure.
from the leaves to the root of the tree. Since one cer-
tain attribute can be the slave of one dependency(tree)
and master for the next dependency(tree) a chain of
transitive dependencies can be created which will be
processed following the constraint propagation pat-
tern (Rossi et al., 2006).
5.5 Results
The OBIK infrastructure is currently being used to
perform comparative benchmarking of hospitals in
Upper Austria. It contains more than 2,000 pa-
tient records including medical parameters like pre-
existing illnesses, risk factors, diagnoses, treatments,
procedures and other administrative data. Talking in
meta model terms, the ontology including data set
consists of 43 entities with 120 attributes and 49 rela-
tions. There are about 40,000 records with more than
188,000 values and about 107,000 references.
First runs of the plausibility checks on the data
yielded tremendous inconsistencies concerning the
semantic data quality - far beyond simple syntactic
and numeric errors; drawbacks that either cause de-
creased usefulness of the statistical outcome or time
and costs on identification and correction. To be more
precise: one of the semantic checks verifies the plau-
sibility of a complication depending on the foregone
medical procedure. It is run at data-input time, which
means the study nurse gets alerted immediately when
she enters implausible data. This check fails in 15
cases out of 2000. Experiences showed that identi-
fication and correction (contacting the study nurse in
the hospital, re-checking the data, ...) takes about 10
minutes on average for each error; resulting in 150
minutes time savings for this check. Currently there
are more than 100 checks implemented, showing sim-
ilar error rates.
AnOntology-basedDataAcquisitionInfrastructure-UsingOntologiestoCreateDomain-independentSoftwareSystems
159

6 CONCLUSIONS
We could show, that ontologies are not only a power-
ful tool for data modeling and knowledge sharing but
also applicable for highly generic software systems.
For this purpose, which was not the origin purpose,
the standard ontology format OWL could not fulfill
all our requirements, which brought us to use a meta
model instead. Aside this format issue, the usage of
ontologies helped to create an absolutely generic data
acquisition and storage system, which can be set up
and maintained by the people who use it, without any
database or programming skills. The intuitive on-
tology editor helps domain experts to manifest their
knowledge into their ontology and the rest of the ap-
plication (data input forms, overview tables, search
and filter functions, export and import interface) are
created automatically on the fly.
REFERENCES
(1997). Meta object facility (mof) specification.
OMG-Document ad/97-08-14.
Astrova, I., Korda, A., and Kalja, A. (2007). Storing owl on-
tologies in sql relational databases. Engineering and
Technology, 29.
Balzert, H. (2010). Java: objektorientiert programmieren:
Vom objektorientierten Analysemodell bis zum objek-
torientierten Programm. W3L-Verl, Herdecke and
Witten, 2 edition.
Baresi, D. L., Nitto, E., and Ghezzi, C. (2006). Toward
open-world software: Issues and challenges. Com-
puter, (06):36–43.
Berners-Lee, T., Hendler, J., and Lassila, O. (2001). The
semantic web a new form of web content that is mean-
ingful to computers will unleash a revolution of new
possibilities.
Chandrasekaran, B., Josephson, J., and Benjamins, V.
(1999). What are ontologies, and why do we need
them? Intelligent Systems and their Applications,
IEEE, 14(1):20–26.
Chen, P. P. S. (1976). The entity relationship model - to-
ward a unified view of data. ACM Transactions on
Database Systems, 1(1):9 – 36.
Ding, Y. (2001). A review of ontologies with the seman-
tic web in view. Journal of Information Science,
27(6):377–384.
Gruber, T. R. (1993). A translation approach to portable on-
tology specifications. Knowl. Acquis., 5(2):199–220.
Horridge, M., Knublauch, H., Rector, A., Stevens, R., and
Wroe, C. (2004). A practical guide to building owl on-
tologies using the protege-owl plugin and co-ode tools
edition 1.0.
Kataria, P., Juric, R., Paurobally, S., and Madani, K. (2008).
Implementation of ontology for intelligent hospital
wards. In Hawaii International Conference on Sys-
tem Sciences, Proceedings of the 41st Annual, ti-
tle=Implementation of Ontology for Intelligent Hos-
pital Wards, page 253.
Kiong, Y. C., Palaniappan, S., and Yahaya, N. A. (2011).
Health ontology system. In Information Technology
in Asia (CITA 11), 2011 7th International Conference
on, pages 1–4.
Knublauch, H., Fergerson, R. W., Noy, N. F., and Musen,
M. A. (2004). 17: The prot
´
eg
´
e owl plugin: An open
development environment for semantic web applica-
tions. In McIlraith, S. ., Plexousakis, D., and van, H.
r. a. n. k., editors, The Semantic Web – ISWC 2004,
volume 3298 of Lecture Notes in Computer Science,
pages 229–243. Springer Berlin / Heidelberg, Berlin
and Heidelberg.
Manola, F. and Mille, E. (2004). Rdf primer, w3c recom-
mendation 10 february 2004.
McGuinness, D. L. and van Harmelen, F. (10 February
2004). Owl web ontology language overview: W3c
recommendation.
Rossi, F., van Beek, P., and Walsh, T. (2006). Handbook
of constraint programming. Elsevier, Amsterdam and
and Boston, 1 edition.
Sirin, E., Parsia, B., Grau, B. C., Kalyanpur, A., and Katz,
Y. (2007). Pellet: A practical owl-dl reasoner. Web
Semantics: Science, Services and Agents on the World
Wide Web, 5(2):51–53.
Smith, B., Williams, J., and Kremer, S. S. (2003). The on-
tology of the gene ontology.
Tran, Q. D. and Kameyama, W. (2007). A proposal
of ontology-based health care information extraction
system: Vnhies. In Research, Innovation and Vision
for the Future, 2007 IEEE International Conference
on, pages 1–7.
Zavaliy, T. and Nikolski, I. (2010). Ontology-based in-
formation system for collecting electronic medical
records data. In Modern Problems of Radio Engineer-
ing, Telecommunications and Computer Science (TC-
SET), 2010 International Conference on, page 125.
KEOD2012-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
160
