
Genetic Algorithms and Firefly Algorithms for Non-linear Bioprocess
Model Parameters Identification
Olympia Roeva
1
and Tanya Trenkova
2
1
Institute of Biophysics and Biomedical Engineering, BAS, 105 Acad. G. Bonchev Str., Sofia 1113, Bulgaria
2
National Institute of Meteorology and Hydrology, BAS, 66 Tzarigradsko shose Bulv., Sofia 1784, Bulgaria
Keywords: Optimization, Genetic Algorithms, Firefly Algorithms, Bioprocess, Identification, Model Parameters.
Abstract: In this paper, Firefly algorithms (FA) and Genetic algorithms (GA) are applied to parameter identification
problem of a non-linear mathematical model of the E. coli cultivation process. A system of ordinary
differential equations is proposed to model the growth of the bacteria, substrate utilization and acetate
formation. Parameter optimization is performed using a real experimental data set from an E. coli MC4110
fed-batch cultivation process. In the considered non-linear mathematical model, the parameters that should
be estimated are maximum specific growth rate, two saturation constants and two yield coefficients.
Parameters of both meta-heuristics are tuned on the basis of several pre-tests according to the optimization
problem considered here. Based on the numerical and simulation result, it is shown that the model obtained
by the FA is more accurate and adequate than the one obtained using the GA. Presented results prove FA
superiority and powerfulness in solving non-linear dynamic model of cultivation processes.
1 INTRODUCTION
Microorganisms have been a subject of particular
attention as a biotechnological instrument, and are
used in so-called cultivation processes. Numerous
useful bacteria, yeasts and fungi are widely found in
nature, but the optimum conditions for growth and
product formation in their natural environment are
seldom discovered.
Cultivation of recombinant microorganisms, e.g.
E. coli, in many cases is the only economical way to
produce pharmaceutic biochemicals such as
interleukins, insulin, interferons, enzymes and
growth factors. Research on E. coli has accelerated
even more since 1997, when its entire genome was
published. Some recent researches and developed
models of E. coli can be found in (Petersen et al.,
2010); (Opalka et al., 2010); (Skandamis and
Nychas, 2000); (Jiang et al., 2010); (Karelina et al.,
2011).
Modelling approaches are central in system
biology and provide new ways towards the analysis
and understanding of cells and organisms. A
common approach to model cellular dynamics is by
using sets of non-linear differential equations. Real
parameter optimization of cellular dynamics models
has become a research field of particularly great
interest. Such problems have widespread
application. The parameter identification of a non-
linear dynamic model is more difficult than that of a
linear one, as no general analytic results exist. The
difficulties that may arise are, for instance,
convergence to local solutions if standard local
methods are used, over-determined models, badly
scaled model function, etc. Due to the non-linearity
and constrained nature of the considered systems,
these problems are very often multimodal. Thus,
traditional gradient-based methods may fail to
identify the good solution. Although a lot of
different global optimization methods exist, the
efficacy of an optimization method is always
problem-specific.
While searching for new, more adequate
modeling metaphors and concepts, methods which
draw their initial inspiration from nature have
received the early attention. During the last decade a
large class of meta-heuristics has been developed
and applied to a variety of areas. The three best
known heuristics are the iterative improvement
algorithms, the probabilistic optimization
algorithms, and the constructive heuristics (Syam
and Al-Harkan, 2010); (Tahouni et al., 2010);
(Brownlee, 2011). Here the attention is focused on
two effective population-based algorithms, namely
Genetic algorithms (GA) and Firefly algorithm (FA).
164
Roeva O. and Trenkova T..
Genetic Algorithms and Firefly Algorithms for Non-linear Bioprocess Model Parameters Identification.
DOI: 10.5220/0004115501640169
In Proceedings of the 4th International Joint Conference on Computational Intelligence (ECTA-2012), pages 164-169
ISBN: 978-989-8565-33-4
Copyright
c
2012 SCITEPRESS (Science and Technology Publications, Lda.)

Holland’s book (Holland, 1992), published in
1975, is generally acknowledged as the beginning of
the research of GA. The GA is a model of machine
learning which derives its behavior from a metaphor
of the processes of evolution in nature (Goldberg,
2006). Since their introduction and subsequent
popularization, the GA have been frequently used as
an alternative optimization tool to the conventional
methods and have been successfully applied to a
variety of areas, and find increasing acceptance
(Akpinar and Bayhan, 2011); (Silva et al., 2009);
(Paplinski, 2010); (Roeva et al., 2010).
The other meta-heuristic algorithm, namely FA,
which idealises some of the flashing characteristics
of fireflies, has been recently developed by Xin-She
Yang (Yang, 2008). According to recent
bibliography, the FA is very efficient and can
outperform other meta-heuristics, such as genetic
algorithms, in solving many optimization problems
(Yang, 2008); (Yang, 2009); (Yang, 2010a; 2010b).
Although the FA has many similarities with other
swarm intelligence based algorithms, it is indeed
much simpler both in concept and implementation
(Yang, 2010a; Yang, 2010b). There are already
several applications of FA to different optimization
problems (Nasiri and Maybodi, 2012);
(Apostolopoulos and Vlachos, 2011); (Yousif et al.,
2011); (Chai-ead et al., 2011). Based on
bibliography results, it is evident that the FA is a
powerful novel population-based method for solving
optimization problems and particularly NP-hard
problems.
In this paper, two optimization algorithms, based
on GA and FA, are proposed for parameter
identification of a fed-batch cultivation process. The
algorithms performances are compared and
analyzed.
2 PROBLEM FORMULATION
There is an increasing interest in technologies that
maximize the production of various essential
enzymes and therapeutic proteins based on E. coli
cultivation. The costs of developing mathematical
models for bioprocesses improvements are often too
high and the benefits are too low. The main reason
for this is related to the intrinsic complexity and
non-linearity of biological systems. The important
part of model building is the choice of a certain
optimization procedure for parameter estimation.
The estimation of model parameters with high
parameter accuracy is essential for successful model
development.
The application of the general state space
dynamical model to the E. coli MC4110 fed-batch
cultivation process leads to the following non-linear
differential equation system (Roeva, 2008):
max
S
dX S F
=XX
dt k S V
(1)
max
/
1
in
SX S
dS S F
=X+S-S
dt Y k S V
(2)
max
/
1
AX A
dA A F
XA
dt Y k A V
(3)
dV
F
dt
(4)
where: X is the biomass concentration, [g·l
-1
]; S is
substrate concentration, [g·l
-1
]; A is acetate
concentration, [g·l
-1
]; F is influent flow rate, [h
-1
]; V
is bioreactor volume, [l];
in
S is influent glucose
concentration, [g·l
-1
]; µ
max
is maximum specific
growth rate, [h
-1
]; Y
S/X
and Y
A/X
are yield coefficients,
[g·g
-1
]; k
S
and k
A
are saturation constants, [g·l
-1
].
The model consists of a set of four differential
Eqs. (1) - (4) thus represented: three dependent state
variables x = [X S A] and five unknown parameters
p = [
max
S
k
A
k
/SX
Y
/AX
Y ].
Parameter estimation problem of the presented
non-linear dynamic system is stated as the
minimization of the distance measure J between the
experimental and the model predicted values of the
considered state variables:
2
exp mod
11
{[ () ()]}
nk
j
ij
J
iimin
yy
(5)
where n is the length of the data vector for each state
variable k; y
exp
are known experimental data; y
mod
are model predictions with a given set of the
parameters.
The cultivation experiments are performed in the
Institute of Technical Chemistry, University of
Hannover, Germany during the collaboration work
with the Institute of Biophysics and Biomedical
Engineering, BAS, Bulgaria, granted by DFG. The
cultivation conditions are presented in details in
Arndt and Hitzmann (2001).
3 FIREFLY ALGORITHM
The Firefly algorithm is a novel meta-heuristic
algorithm which is inspired from flashing light
GeneticAlgorithmsandFireflyAlgorithmsforNon-linearBioprocessModelParametersIdentification
165

behaviour of fireflies in nature. Based on Yang
(2008) the basic steps of the FA can be summarized
as the following pseudo code:
begin
Define light absorption coefficient γ
initial attractiveness β
0
randomization parameter α
objective function f(x), where
x = (x
1
, ..., x
d
)
T
Generate initial population of
fireflies x
i
(i = 1, 2, ..., n)
Determine light intensity I
i
via f(x
i
)
while (t < MaxGeneration) do
for i = 1 : n all n fireflies do
for j = 1 : i all n fireflies do
if (I
j
> I
i
) then
Move firefly i towards j
based on Eq. (8)
end if
Attractiveness varies with
distance r via exp[−γr
2
]
Evaluate new solutions and
update light intensity
end for j
end for i
Rank the fireflies and find
the current best
end while
Postprocess results and visualization
end begin
For simplicity, it is assumed that the attractiveness
of a firefly is determined by its brightness, which in
turn is associated with the encoded objective
function of the optimization problems.
Attractiveness. In FA, each firefly has a location
x = (x
1
, ..., x
d
)
T
in a d-dimensional space and light
intensity I(x) or attractiveness β(x), which are
proportional to an objective function f(x).
Attractiveness β(x) and light intensity I(x) are
relative and these should be judged by the rest
fireflies. Thus, attractiveness will vary with the
distance r
ij
between firefly i and firefly j. So
attractiveness β of a firefly can be defined by Eq. (6)
(Yang, 2009); (Yang, 2010a; 2010b):
0
()
m
r
re
, m ≥ 1
(6)
where r or r
ij
is the distance between the i-th and j-th
of two fireflies. β
0
is the initial attractiveness at r = 0
and γ is a fixed light absorption coefficient that
controls the decrease of the light intensity. In the
herewith applied FA m = 2.
Distance and movement. The initial solution is
generated based on
x
j
= rand*(Ub − Lb) +Lb (7)
where rand is a random number generator uniformly
distributed in the space [0, 1]; Ub and Lb are the
upper range and lower range of the j-th firefly
(variable), respectively.
When firefly i is attracted to another more
attractive (brighter) firefly j, its movement is
determined by:
2
10
1
()( )
2
ij
r
ii ij
x x e x x rand
(8)
where the first term is the current position of a
firefly, the second term is used for considering a
firefly's attractiveness to light intensity seen by
adjacent fireflies β(r) (Eq. (6)), and the third term is
used to describe the random movement of a firefly in
case there are no brighter ones. The coefficient α is a
randomization parameter determined by the problem
of interest. The distance r
i,j
between any two fireflies
i and j at x
i
and x
j
, respectively, is defined as a
Cartesian or Euclidean distance (Yang, 2009):
2
,,
1
()
d
ij i j i k j k
k
rxx xx
(9)
where x
i,k
is the k-th component of the spatial
coordinate x
i
of the i-th firefly.
4 GENETIC ALGORITHM
A pseudo code of a GA is presented as:
begin
i = 0
Generate initial population P(0)
Evaluate P(0) fitness
while (t < MaxGeneration) do
for i = 1 : n all n chromosomes do
Select P(i) from P(i
– 1)
Recombine P(i) with probability p
C
Mutate P(i) with probability p
m
Evaluate P(i) fitness
end for
end while
Rank the chromosomes, find
the current best and save
end begin
Solution Representation. Each individual or
chromosome is made up of a sequence of genes from
a certain alphabet. Binary representation is the most
common one, mainly because of its relative
simplicity. A binary 20-bit representation is
considered here. Five model parameters are
represented in the chromosome – maximum specific
growth rate (
max
), two saturation constants (k
S
and
IJCCI2012-InternationalJointConferenceonComputationalIntelligence
166

k
A
), and two yield coefficients (Y
S/X
and Y
A/X
). The
following upper and lower bounds are considered:
0 <
max
< 0.8; 0 < k
s
< 1; 0 < k
A
, Y
S/X
, Y
A/X
< 30.
Selection Function. The selection method used here
is the roulette wheel selection. The probability P
i
for
each individual is defined by:
1
i
i
PopSize
j
j
F
P
F
,
(10)
where F
i
equals the fitness of individual i and
PopSize is the population size.
Genetic Operators. There are two basic types of
operators: crossover and mutation. Let
X
and
Y
be
two
m-dimensional row vectors denoting parents
from the population. For
X
and Y binary, binary
mutation and simple crossover are defined:
1,if (0,1)
, otherwis
e
im
i
i
x
Up
x
x
(11)
,if ,if
,
, otherwise , other
wise
ii
ii
ii
x
ir y ir
xy
yx
,
(12)
where
p
m
is the probability of binary mutation, r is a
random number from a uniform distribution from 1
to
m.
Initialization, Termination and Evaluation
Functions.
GA must provide an initial population.
The most common method is to randomly generate
solutions for the entire population. The GA moves
from generation to generation selecting and
reproducing parents until a termination criterion is
met. The most frequently used stopping criterion is a
specified maximum number of generations.
Evaluation functions of many forms can be used in a
GA, subject to the minimal requirement that the
function can map the population into a partially
ordered set. As stated, the evaluation function is
independent of the GA.
5 RESULTS AND DISCUSSION
A series of parameter identification procedures for
the considered model Eq. (1) - (4), using FA and
GA, are performed. The computer specifications to
run all optimization procedures are Intel® Core™i5-
2320 CPU @ 3.00GHz, 8 GB Memory (RAM),
Windows 7 (64bit) operating system.
Each algorithm has its own influential
parameters that affect its performance in terms of
solution quality and computational time. In order to
increase the performance of the FA and GA, it is
necessary to provide the adjustments of the
parameters depending on the problem domain. With
the appropriate choice of the algorithm settings the
accuracy of the decisions and the execution time can
be optimized. Parameters of the FA are tuned on the
basis of a large number of pre-tests according to the
parameter identification problem, considered here.
After tuning procedures the main FA parameters are
set to the optimal settings (see Table 1).
Table 1: Firefly algorithm parameters.
Firefly algorithm parameter Value
Attractiveness, β
0
1
light absorption coefficient, γ 1
randomization parameter, α 0.2
number of fireflies 60
number of iterations 100
In Table 2, the GA parameters used in this work
are presented. These settings are chosen on the basis
of performed pre-test procedures and the results in
(Roeva, 2008). For fair and realistic comparison, the
GA is run for the same number of function
evaluations (
N
FE
) of FA − 1200.
Table 2: Genetic algorithm parameters.
Genetic algorithm parameter Value
generation gap 0.97
crossover rate 0.70
mutation rate 0.05
precision of binary representation 20
number of individuals 60
number of generations 100
Because of the stochastic characteristics of the
applied algorithm, FA and GA have been run at least
30 times in order to carry out meaningful statistical
analysis. The mean results of the parameters
estimates, total time for the solver to run (
T) and
objective function value
J (Eq. (5)) are observed.
The obtained results are summarized in Table 3. The
obtained results from both population-based
algorithms are very close. But if the results are
scrutinized more carefully, it is evident that for 1200
function evaluations the GA obtained worse results
compared to the FA performance. For the same
computational time and the same number of function
evaluations the FA obtained
J = 6.03, while GA – J
= 6.20. A graphical representation of the
convergence of the objective function
J for both
algorithms with time (iterations) is shown (in
logarithmic scale) in Fig. 1.
GeneticAlgorithmsandFireflyAlgorithmsforNon-linearBioprocessModelParametersIdentification
167
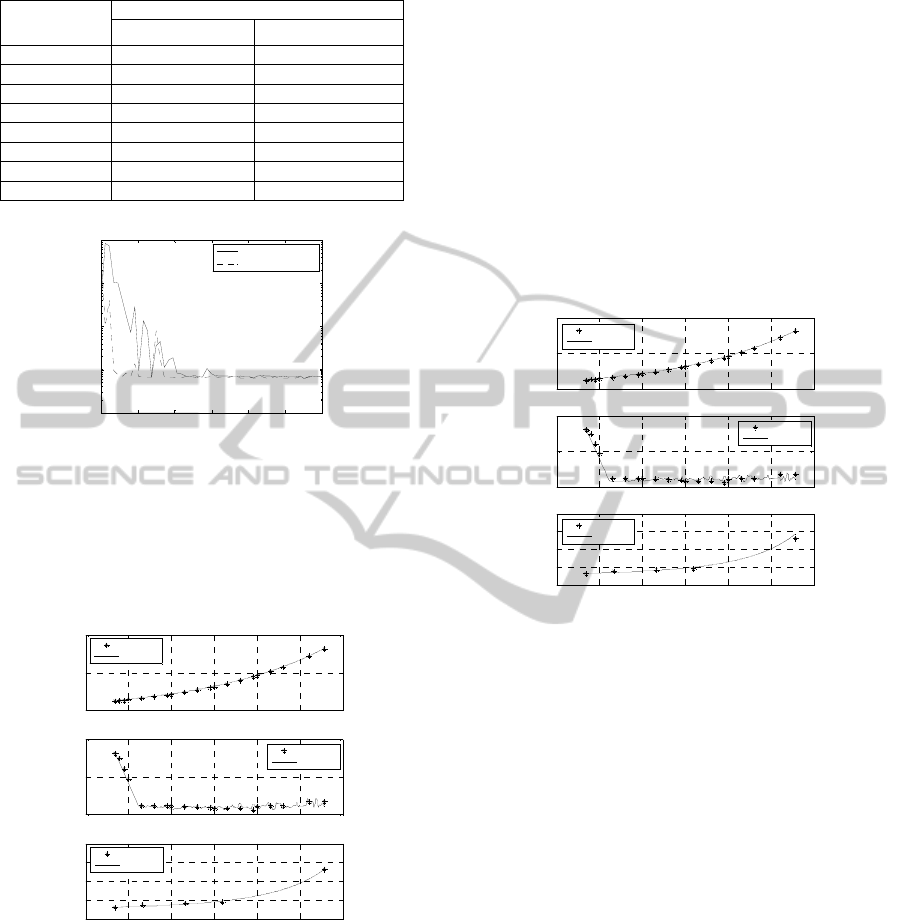
Table 3: Identified model parameters.
Model
parameters
Estimated values
Firefly algorithm Genetic algorithm
µ
max
0.4663 0.4723
k
S
0.0129 0.0139
k
A
5.4416 4.5161
Y
S/
X
2.0099 2.0104
Y
A
/
X
29.2083 24.1935
J
6.0259 6.2007
T 131.9561 132.5072
N
FE
1200 1200
0 1000 2000 3000 4000 5000 6000
10
0
10
1
10
2
10
3
10
4
Objective function through iterations
Iterations
Objective function
Genetic Algorithm
Firefly Algorithm
Figure 1: Convergence of the objective function with time.
The FA algorithm shows better convergence
performance in the beginning of the optimization
process, compared to the GA. The FA converges
faster than the GA and achieves lower value for
J in
the end of the optimization.
6 7 8 9 10 11 12
0
5
10
Results from Firefly Algorithm
Biomass, [g/l]
exp. data
model data
6 7 8 9 10 11 12
0
0.5
1
Substrate, [g/l]
exp. data
model data
6 7 8 9 10 11 12
0
0.05
0.1
0.15
0.2
Time, [h]
Acetate, [g/l]
exp. data
model data
Figure 2: Time profiles of the process variables:
experimental data and models predicted data – FA result.
In the next two figures the modelled E. coli fed-
batch cultivation process variables (biomass,
substrate and acetate) and the measured ones (real
experimental data) are presented. In most cases,
graphical comparisons clearly show the existence or
absence of systematic deviations between model
predictions and measurements. It is evident that a
quantitative measure of the differences between
calculated and measured values is an important
criterion for the adequacy of a model. Figs. 2 and 3
show that there is a coincidence between the
measured estimates and those modelled with both
algorithms.
Hence, the difference between the values of the
objective function achieved by FA and GA comes
mainly from the value of the substrate and is
negligible from the value of the acetate, achieved by
them. As it can be seen from Fig. 2, the model
obtained on the basis of FA predicts more accurately
the substrate and acetate dynamics in comparison to
the GA model (Fig. 3). Thus, the presented results
show that the FA is more powerful in solving the
optimization problem, considered here.
6 7 8 9 10 11 12
0
5
10
Biomass, [g/l]
Results from Genetic Algorithm
exp. data
model data
6 7 8 9 10 11 12
0
0.5
1
Substrate, [g/l]
exp. data
model data
6 7 8 9 10 11 12
0
0.05
0.1
0.15
0.2
Time, [h]
Acetate, [g/l]
exp. data
model data
Figure 3: Time profiles of the process variables:
experimental data and models predicted data – GA result.
6 CONCLUSIONS
The Firefly algorithm, recently developed by Yang
(2008), is a very powerful novel population-based
method. The social behavior and the flashing light of
fireflies can be easily associated with the objective
function of a given optimization problem. In this
paper, FA is proposed and tested for application to
the parameter identification of a non-linear
dynamical model of
E. coli cultivation process. A
comparison of Firefly algorithm and Genetic
algorithm is done. The mathematical model is
considered as a system of four ordinary differential
equations, describing the three considered process
variables
– biomass, substrate and acetate
concentrations. Numerical and simulation results
from model parameter identification based on FA
and GA reveal that correct and consistent results can
be obtained using the discussed meta-heuristics. The
algorithms comparison shows that the model
obtained by means of the FA is more accurate and
adequate than the one based on GA. Finally, the
IJCCI2012-InternationalJointConferenceonComputationalIntelligence
168

results confirm that the Firefly algorithm is powerful
and efficient tool for identification of the parameters
in the bioprocess model parameter optimization
problem.
ACKNOWLEDGEMENTS
The investigations are partially supported by the
Bulgarian National Science Fund, Grants DID 02/29
and DMU 02/4.
REFERENCES
Apostolopoulos, T. and Vlachos, A., (2011). Application
of the Firefly Algorithm for Solving the Economic
Emissions Load Dispatch Problem. International
Journal of Combinatorics, Article ID 523806.
Akpinar, S. and Bayhan, G. M., (2011). A Hybrid Genetic
Aalgorithm for Mixed Model Assembly Line
Balancing Problem with Parallel Workstations and
Zoning Constraints. Engineering Applications of
Artificial Intelligence, 24(3), 449-457.
Arndt, M. and Hitzmann, B., (2001). Feed
Forward/feedback Control of Glucose Concentration
during Cultivation of Escherichia coli. 8th IFAC Int
Conf on Comp Appl in Biotechn, Canada, 425-429.
Chai-ead, N., Aungkulanon, P., Luangpaiboon, P., (2011).
Bees and firefly algorithms for noisy non-linear
optimisation problems. Prof. Int. Multiconference of
Engineers and Computer Scientists, 2, 1449-1454.
Silva, F., Sánchez Pérez, J. M., Gómez Pulido, J. A.,
Vega-Rodríguez, M. A., (2009). AlineaGA - A
Genetic Algorithm with Local Search Optimization for
Multiple Sequence Alignment. Applied Intelligence,
32(2), Springer, Berlin Heidelberg, 164-172.
Goldberg, D. E., (2006). Genetic Algorithms in Search,
Optimization and Machine Learning. Addison Wesley
Longman, London.
Holland, J. H., (1992). Adaptation in Natural and
Artificial Systems (2nd ed.). Cambridge, MIT Press.
Jiang, L., Ouyang, Q., Tu, Y., (2010). Quantitative
Modeling of Escherichia coli Chemotactic Motion in
Environments Varying in Space and Time. PLoS
Comput Biol, 6(4), e1000735. doi:10.1371/
journal.pcbi.1000735.
Karelina, T. A., Ma, H., Goryanin, I., Demin, O. V.,
(2011). EI of the Phosphotransferase System of
Escherichia coli: Mathematical Modeling Approach to
Analysis of Its Kinetic Properties. J of Biophysics,
Article ID 579402, doi:10.1155/2011/579402.
Nasiri, B. and Meybodi, M. R., (2012). Speciation-based
firefly algorithm for optimization in dynamic
environments. Int J Artificial Intelligence, 8(S12),
118-132.
Opalka, N., Brown, J., Lane, W. J., Twist, K.-A. F.,
Landick, R., Asturias, F. J., Darst, S. A., (2010).
Complete Structural Model of Escherichia coli RNA
Polymerase from a Hybrid Approach. PLoS Biol, 8(9),
e1000483. doi:10.1371/journal.pbio.1000483.
Paplinski, J. P., (2010). The Genetic Algorithm with
Simplex Crossover for Identification of Time Delays.
Intelligent Information Systems, 337-346.
Petersen, C. M., Rifai, H. S., Villarreal, G. C., Stein, R.,
(2011). Modeling Escherichia coli and Its Sources in
an Urban Bayou with Hydrologic Simulation Program
-- FORTRAN, Journal of Environmental Engineering.
137(6), 487-503.
Roeva, O., (2008). Parameter Estimation of a Monod-type
Model based on Genetic Algorithms and Sensitivity
Analysis. LNCS, Springer-Verlag Berlin Heidelberg,
4818, 601-608.
Roeva, O., Kosev, K., Trenkova, T., (2010). A modified
multi-population genetic algorithm for parameter
identification of cultivation process models. IJCCI
(ICEC) 2010, Valencia, Spain, 348-351.
Skandamis, P. N. and Nychas, G. E., (2000). Development
and Evaluation of a Model Predicting the Survival of
Escherichia coli O157:H7 NCTC 12900 in
Homemade Eggplant Salad at Various Temperatures,
pHs, and Oregano Essential Oil Concentrations. AEM,
66(4), 1646-1653.
Syam, W. P. and Al-Harkan, I. M., (2010). Comparison of
Three Meta Heuristics to Optimize Hybrid Flow Shop
Scheduling Problem with Parallel Machines. WASET,
62, 271-278.
Tahouni, N., Smith, R., Panjeshahi, M. H., (2010).
Comparison of Stochastic Methods with Respect to
Performance and Reliability of Low-temperature Gas
Separation Processes. The Canadian Journal of
Chemical Engineering, 88(2), 256-267.
Yang, X. S., (2008). Nature-Inspired Meta-Heuristic
Algorithms, Luniver Press, Beckington, UK.
Yang, X. S., (2009). Firefly algorithm for multimodal
optimization, LNCS, Springer-Verlag Berlin
Heidelberg, 5792, 169-178.
Yang, X. S., (2010a). Firefly algorithm, stochastic test
functions and design optimisation, International
Journal of Bio-Inspired Computation, 2(2), 78-84.
Yang, X. S., (2010b). Firefly algorithm, Levy flights and
global optimization, Research and Development in
Intelligent Systems XXVI, Springer, London, UK, 209-
218.
Yousif, A., Abdullah, A. H., Nor, S. M., Abdelaziz, A. A.,
(2011). Scheduling Jobs on Grid Computing Using
Firefly Algorithm, Journal of Theoretical and Applied
Information Technology, 33(2), 155-164.
GeneticAlgorithmsandFireflyAlgorithmsforNon-linearBioprocessModelParametersIdentification
169
