
Structure Prediction with FAMS for Proteins Screened Critically
to Autoimmune Diseases based upon Bioinformatics
Shigeharu Ishida
1
, Hideaki Umeyama
2
, Mitsuo Iwadate
2
and Y-h. Taguchi
1
1
Department of Physics, Chuo University, Tokyo 112-8551, Japan
2
Department of Biological Sciences, Chuo University, Tokyo 112-8551, Japan
Keywords:
FAMS, Autoimmune Diseases, Structure Prediction, Drug Discovery.
Abstract:
Drug discovery for autoimmune diseases is recently recognized to be an important task. In this study, we try to
perform structure prediction of proteins whose gene promoter regions were previously reported to be specif-
ically methelysed or demethylased commonly for three autoimmune diseases, systemic lupus erythematosus,
rheumatoid arthritis, and dermatomyositis. FAMS were employed for this purpose and we can predict three
dimensional structure with significantly small enough P-values. Most of them are suggested to be self im-
munology related proteins and will be important drug target candidates. We also found some proteins which
form complex with each other. The possibility of a new drug target, i.e., suppression of protein complex
formation is suggested.
1 INTRODUCTION
Autoimmune diseases are recently recognized as seri-
ous symptom. For example, systemic lupus erythe-
matosus (SLE), which is known to be one of sys-
temic autoimmune diseases, most often harms the
heart, joints, skin, lungs, blood vessels, liver, kidneys,
and nervous system. The cause of this disease is un-
known. The lack of the knowledge about basic mech-
anism of the disease prevents us from generating ef-
fective drugs to cure this disease. SLE is the secondly
frequent connectivetissue disease, while the most fre-
quent one is Rheumatoid Arthritis (RA), which is also
known to be one of autoimmune diseases. Although
there are some proposals about the cause of RA, it has
not yet been fully understood. In RA, the arthritis of
joints known as synovitis is inflammation of the syn-
ovial membrane that lines joints and tendon sheaths.
Joints become swollen, tender and warm, and stiff-
ness limits their movement. Another example of au-
toimmune disease is dermatomyositis (DM), which is
also a connective-tissue disease related to polymyosi-
tis that is characterized by inflammation of the mus-
cles and the skin. Its cause is unknown, too.
In spite of the lack of basic understanding of dis-
eases’ causes, there is a general belief; there should be
a common cause of autoimmune diseases. Following
this line, in accordance with the recent development
of genome science, several conjectures are proposed.
For example, O’Hanlon et al recently showed that
there are common pathways which contribute to
multiple systematic autoimmune diseases (O’Hanlon
et al., 2011b), based upon gene expression analy-
sis. More recently, they have confirmed their findings
using proteomic analysis (O’Hanlon et al., 2011a).
However, Zhou et al (Zhou et al., 2005) found that
unaffected monozygotic (MZ) twins share fibroblast
gene expression with systemic sclerosis (SSc) pa-
tients (counter parts). SSc is also believed to be re-
lated to autoimmune diseases. On the other hand,
Gervin et al (Gervin et al., 2012) recently found
that combined analysis between gene expression and
methylation enables them to detect slight difference
between affected and unaffected twins. Their find-
ings are not contradict to the study by Javierre et
al (Javierre et al., 2010) who could not find any
shared methylation patterns among multiple autoim-
mune diseases. Thus, at the moment, it is a little bit
confusing what kind of aspects can be shared with
multiple autoimmune diseases.
A few years ago, we reanalyzed (Taguchi, 2010)
Javierre et al’s data (Javierre et al., 2010) using prin-
cipal component analysis (PCA) and found that some
genes’ methylation are commonly and significantly
different from healthy controls. In this paper, we try
to validate our findings using Full Automatic Model-
ing System (FAMS), which is protein structure pre-
diction program that perform comparative modeling
261
Ishida S., Umeyama H., Iwadate M. and Taguchi Y..
Structure Prediction with FAMS for Proteins Screened Critically to Autoimmune Diseases based upon Bioinformatics.
DOI: 10.5220/0004188802610267
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2013), pages 261-267
ISBN: 978-989-8565-35-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

with database search and simulated annealing. Using
FAMS(Umeyama and Iwadate, 2002), we can predict
functionality of genes by comparing them with the
proteins whose function and structure are known. We
also validate if these genes can form complex and find
many candidates to form protein complex. The possi-
bility that they can be a drug target will be discussed.
2 MATERIALS AND METHODS
2.1 Selection of Candidate Genes
Although details are reported previously(Taguchi,
2010), here we briefly describe how we have selected
candidate genes. Javierre et al (Javierre et al., 2010)
measured promoter methylation patterns using mi-
croarray technology (Illumina GoldenGate Methyla-
tion Cancer Panel I) for SLE, RA, and DM. Their
expression patterns are deposited to Gene Expression
Omnibus with the accession number of GSE19033.
We downloaded series
matrix.txt from there, applied
PCA to them and picked up gene whose promot-
ers’ methylation is significantly different from healthy
controls.
2.2 Structure Prediction of Selected
Genes
Selected genes’ amino acid sequences are down-
loaded from SWISS Prot. Then their protein struc-
tures are inferred by FAMS.
2.3 Protein Complex Formation
Prediction
We checked if a pair of model proteins used for struc-
ture prediction can form protein complex or not as
follows. First, PDB files which contains at least
one model protein as a member of protein com-
plex are downloaded. Then, which model proteins
are included into the common PDB files is investi-
gated. Thirdly, inter-atomic distances between pairs
of model proteins which belong to the same PDB file
are computed. If there are at least a pair of atoms
whose distances are less than 3.5
˚
A, a pair of model
proteins is listed as a candidate to form protein com-
plex.
Figure 1: Comparison between reference protein
2OQ0(Green) and model protein AIM2(Cyan).
3 RESULTS
3.1 Biological Significance Figured out
by FAMS
In Table 1, we have listed genes (i.e., reference pro-
teins) selected by PCA(Taguchi, 2010), together with
the model proteins which are inferred to have sim-
ilar structure to each of them by FAMS. First of
all, FAMS has successfully listed model proteins for
most of reference proteins with very small P-values.
Fig. 1 shows a typical example of model proteins.
It is the model protein 2OQ0
B for the reference
protein AIM2. Alignment regions are 192 amino
acid sequence from total length of 209 amino acid
of 2OQ0
B and 191 amino acid sequence from to-
tal length 343 amino acid sequence of AIM2. Se-
quence similarity between two alignment regions is
44 %. P-value attributed is 4× 10
−92
. Although this
is only one example of typical relationship between
model/reference proteins, generally we could get this
quality of structural similarities. This means struc-
tural similarity between models and references is reli-
able. In addition to this, biological features attributed
to the model proteins are often reasonable. Due to the
limitation of the space, we cannot explain all of them
one by one, we will point out some of these exam-
ples. Then, modeling yields predictions that need to
be experimentally verified.
TRIP6 is expected to have similar structure to
CRP1, which is inferred as immune response(Latonen
et al., 2010). TM7SF3 is recognized as cytochrome
c oxidase, which was reported to bind to immune
gamma-globulins (Frey et al., 1978). TIE1, PECAM1
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
262
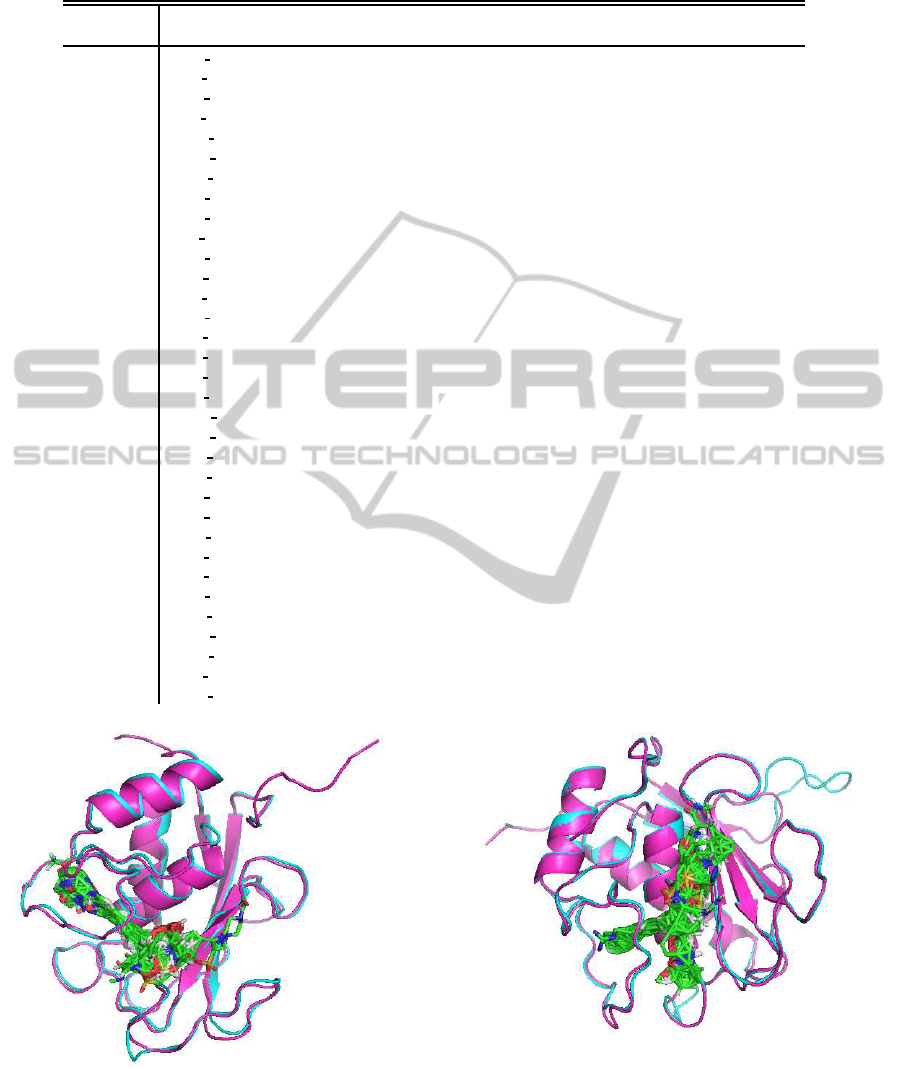
Table 1: Selected genes and model protein used for structure prediction. Bold ID of PDB indicates that reference protein itself
is detected in PDB.
Reference Model
gene symbol
PDB ID P-value gene symbol
AIM2 2OQ0 B 4× 10
−92
GAMMA-INTERFERON-INDUCIBLE PROTEIN IFI-16
CARD15
3CIY B 7× 10
−64
TOLL-LIKE RECEPTOR 4, VARIABLE LYMPHOCYTE (TLR4)
CD82
2BG9 A 0.46 ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA CHAIN
CSF1R
3B43 A 5× 10
−83
TITIN
CSF3
1GNC A 2× 10
−66
GRANULOCYTE COLONY-STIMULATING FACTOR
CSF3R
3DMK A 1× 10
−71
DOWN SYNDROME CELL ADHESION MOLECULE (DSCAM)
DHCR24
2Q4W A 1× 10
−115
CYTOKININ DEHYDROGENASE 7 (CKO7)
ERCC3
2W74 D 1× 10
−152
TYPE I RESTRICTION ENZYME ECOR124II R PROTEIN (HSDR)
GRB7
3HK0 B 2× 10
−73
GROWTH FACTOR RECEPTOR-BOUND PROTEIN 10 (GRB10)
HGF
2F83 A 1× 10
−111
COAGULATION FACTOR XI
HOXB2
2D5V A 9× 10
−24
HEPATOCYTE NUCLEAR FACTOR 6 (HNF-6)
IFNGR2
1FNF A 1× 10
−37
FIBRONECTIN
LCN2
1X71 A 1× 10
−51
NEUTROPHIL GELATINASE-ASSOCIATED LIPOCALIN (NGAL)
LMO2
2XJY A 2× 10
−33
RHOMBOTIN-2
LTB4R
2KS9 A 2×10
−83
SUBSTANCE-P RECEPTOR
MMP14
1SU3 B 1×10
−160
INTERSTITIAL COLLAGENASE (MMP-1)
MMP8
1SU3 B 1×10
−171
INTERSTITIAL COLLAGENASE (MMP-1)
MPL
3L5H A 4× 10
−63
INTERLEUKIN-6 RECEPTOR SUBUNIT BETA (IL6RB)
PAD14
2DEW X 0.0 PROTEIN-ARGININE DEIMINASE TYPE IV
PECAM1
3DMK A 1× 10
−104
DOWN SYNDROME CELL ADHESION MOLECULE (DSCAM)
PI3
1TWP A 2×10
−19
WHEY ACIDIC PROTEIN (WAP)
RARA
3DZY A 4× 10
−95
RETINOIC ACID RECEPTOR RXR-ALPHA
S100A2
2RGI A 4× 10
−19
PROTEIN S100-A2
SEPT9
3FTQ A 1× 10
−137
SEPTIN-2
SLC22A18
1PW4 A 1× 10
−108
GLYCEROL-3-PHOSPHATE TRANSPORTER
SPI1
1GVJ B 1×10
−21
C-ETS-1 PROTEIN (ETS1)
SPP1
1D2T A 3× 10
−14
ACID PHOSPHATASE (ACP)
STAT5A
1Y1U A 0.0 SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION (STAT5A)
SYK
2OZO A 1× 10
−168
TYROSINE-PROTEIN KINASE ZAP-70
TIE1
3DMK A 2× 10
−84
DOWN SYNDROME CELL ADHESION MOLECULE (DSCAM)
TM7SF3
[1AR1 A] 6× 10
−88
CYTOCHROME C OXIDASE
TRIP6
1B8T A 2×10
−32
CYSTEINE-RICH PROTEIN 1 (CRP1)
VAMP8
2KOG A 1× 10
−21
VESICLE-ASSOCIATED MEMBRANE PROTEIN 2 (VAMP2)
Figure 2: Ligand binding to MMP8. Magenta and Cyan
are reference and model proteins. Stick models are ligand
molecules binding to 95% homologous proteins with refer-
ence.
Figure 3: Ligand binding to MMP14. Magenta and Cyan
are reference and model proteins. Stick models are ligand
molecules binding to 95% homologous proteins with refer-
ence.
StructurePredictionwithFAMSforProteinsScreenedCriticallytoAutoimmuneDiseasesbaseduponBioinformatics
263

Table 2: The number of common PDB ID detected homology search results between two genes are listed. The threshold of
both searches set to 1× 10
−10
, that is enough low to conserve the protein tertiary structure. Then the model proteins is likely
to bind to other model proteins.
AIM2 P624 F
CARD15
P302 R
CD82
P557 R
CSF1R E26 F
CSF3
E242 R
CSF3R P472 F
DHCR24
P652 R
ERCC3
P1210 R
GRB7
E71 R
HGF
E102 R
HOXB2
P99 F
IFNGR2
P377 R
LCN2
P86 R
LMO2
E148 F
LTB4R P163 F
MMP14 P13 F
MMP8
E89 R
MPL
P62 F
PADI4 E24 F
PECAM1
E32 R
PI3
P274 R
RARA
P1076 R
S100A2
E36 R
SEPT9 P374 F
SLC22A18
P216 R
SPI1
E205 F
SPP1 P647 F
STAT5A
P704 R
SYK P584 F
TIE1 E66 R
TM7SF3
P1068 R
TRIP6
P1090 F
VAMP8 P241 F
AIM2 P624 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
CARD15
P302 R 0 0 2 0 5 0 16 0 3 0 0 0 0 0 0 0 0 0 5 0 0 0 0 0 0 0 8 0 0 0 0 0
CD82
P557 R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
CSF1R
E26 F 0 2 0 0 186 0 0 10 25 0 5 8 0 0 0 0 5 0 410 0 0 2 0 0 0 0 9 1223 1355 0 0 0
CSF3
E242 R 0 0 0 0 52 0 0 0 0 0 42 0 0 0 0 0 28 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0
CSF3R
P472 F 0 5 0 186 52 0 0 0 107 0 166 48 0 3 0 0 115 0 1177 0 0 0 0 0 0 0 0 0 204 0 0 0
DHCR24
P652 R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
ERCC3
P1210 R 0 16 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
GRB7
E71 R 0 0 0 10 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 170 211 10 0 0 0
HGF
E102 R 0 2 0 25 0 107 0 0 0 0 140 2 0 0 0 0 0 0 84 2 0 0 0 0 0 0 0 0 0 0 0 0
HOXB2
P99 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
IFNGR2
P377 R 0 0 0 5 42 166 0 0 0 140 0 0 0 0 0 0 91 0 24 0 0 0 0 0 0 0 0 0 54 0 0 0
LCN2
P86 R 0 0 0 4 0 24 0 0 0 2 0 0 0 0 0 0 0 0 16 0 0 0 0 0 0 0 0 0 0 0 0 0
LMO2
E148 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0
LTB4R
P163 F 0 0 0 0 0 3 0 0 0 0 0 0 0 0 0 0 0 0 6 0 0 0 0 0 0 0 0 0 0 0 0 0
MMP14
P13 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 125 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
MMP8
E89 R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 125 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
MPL
P62 F 0 0 0 5 28 115 0 0 0 0 0 91 0 0 0 0 0 0 19 0 3 0 0 0 0 0 0 0 34 0 0 0
PADI4
E24 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
PECAM1
E32 R 0 5 0 410 1 1177 0 0 0 84 0 24 32 0 6 0 0 19 0 0 0 0 0 0 0 0 0 1 129 0 0 0
PI3
P274 R 0 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
RARA
P1076 R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0
S100A2
E36 R 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 2 1 1 0 0 0
SEPT9
P374 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0
SLC22A18
P216 R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
SPI1
E205 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
SPP1
P647 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
STAT5A
P704 R 0 8 0 9 0 0 0 0 170 0 0 0 0 0 0 0 0 0 0 0 0 0 2 1 0 0 0 164 9 0 0 0
SYK
P584 F 0 0 0 1223 0 0 0 0 211 0 0 0 0 0 0 0 0 0 0 1 0 0 1 0 0 0 0 164 1200 0 0 0
TIE1
E66 R 0 0 0 1355 0 204 0 0 10 0 0 54 0 0 0 0 0 34 0 129 0 0 1 0 0 0 0 9 1200 0 0 0
TM7SF3
P1068 R 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
TRIP6
P1090 F 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
VAMP8
P241 F 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
and CSF3R are recognized as DOWN SYNDROME
CELL ADHESION MOLECULE (DSCAM), which
is known to be immunoglobulin (Ig)-superfamily re-
ceptor in insect(Watson et al., 2005). SYK is rec-
ognized as TYROSINE-PROTEIN KINASE ZAP-70
and both SYK and ZAP-70 are reported to display dis-
tinct requirements for Src family kinases in immune
response receptor signal transduction(Zoller et al.,
1997). STAT5A itself is found in PDB, which is re-
ported to play critical role for cytokine responses and
normal immune function(Lin et al., 2012). SPI1 is
recognized as ETS1, which is known to be expres-
sive in SLE and play some function in immune sys-
tem(Pan et al., 2011). S100-A2 itself is in PDB and is
reported to be antibodies and inhibitors directed to-
ward receptor for advanced glycation end products
(RAGE) ligands(Heijmans et al., 2012). RARA is
structurally similar to RXR-α, which is reported to
be involved in inflammatory responses(Selvaraj et al.,
2010). PI3 is as WAP, which is reported to play a
role in innate immune(Bingle and Vyakarnam, 2008).
PADI4 is itself in PDB and is reported to be important
in RA(Abd-Allah et al., 2012). MPL’s structure is in-
ferred to be similar to IL6RB. IL6R is reported to be a
key mediator of RA(Cronstein, 2007). LCN2, which
is also called as NGAL, is in PDB. NGAL is tried to
be used as a marker of inflammatory status for allow-
ing an early diagnosis of inflammatory disease such
as autoimmune disease in DS patients(Dogliotti et al.,
2010). One of HOXB2’s model proteins is HNF-
6, which is known to cause immunologically distinct
feature(Samadani and Costa, 1996). AIM2 is struc-
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
264

Figure 4: Protein complex formation candidates between
PECAM1 (Cyan) and CSF1R (Magenta) detected in P-
value 1e-53 and 4e-68, based upon protein structure of PDB
2ZJS. Green region are excluded from matching between
model and reference proteins.
turally similar to IFI16. AIM2 and IFI16 are reported
to play critucal role in immunology (Jin et al., 2012).
CARD15 is inferred to be similar to TLR4 which play
a role in cell antiviral response together with TLR3:
TICAM1-specific signaling pathways(Meylan et al.,
2004). CD82 is known to be ACETYLCHOLINE
RECEPTOR PROTEIN which often play a critical
role in immune system(Quek et al., 2012)
1
. CSF1R
is assigned to be TITAN, which is known to be in-
volved in to immune response(Skeie et al., 1998).
SPP1 is recognized as ACID PHOSPHATASE, which
is known to be related to be autoimmune prostati-
tis(Fong et al., 1997). LMO2, which is also known to
be RHOMBOTIN-2, is known to be related to ZFAT
(a zinc-finger gene in autoimmune thyroid disease
susceptibility region / an immune-related transcrip-
tional regulator containing 18 C2H2-type zinc-finger
domains and one AT-hook)(Tsunoda et al., 2010).
DHCR24 is regarded as CYTOKININ DEHYDRO-
GENASE. Cytokine has, not to mention, been used
to refer to the immunomodulating agents. SEPT9 is
homologous to SEPTIN-2, which is reported to be up-
regulated in cytoskeletal and immune function-related
proteome profiles (Gabr et al., 2007). IFNGR2 is re-
garded as FIBRONECTIN, which play a role in im-
mune responses in organ transplant recipients(Coito
et al., 2000). CSF3 itself is in PDB, which is knownto
have relationship with immune system (Sarkar et al.,
2012). GRB7 is also recognized as GRB10, which
play an important role in immune system, although it
is in cancer(O-Sullivan et al., 2008). HGF is related to
COAGULATION FACTOR XI, which is known to be
related to immunology(Bouma et al., 1983). LTB4R
1
Although P-value attributed to CD82 is not small
enough, reliability of this assignment turns out to be rea-
sonable after some more details consideration (not shown
here).
is recognized as SUBSTANCE-P RECEPTOR, which
is known to have immune response to respiratory syn-
cytial virus infection (Tripp et al., 2002).
These are only a part of immune system related
features which are attributed to each gene by FAMS.
Although more examples can easily be listed, we omit
the rest of them because of length limitation. Anyway,
it is clear that FAMS based feature attribution works
very well for genes selected by PCA(Taguchi, 2010).
3.2 Possibility of Drug Discovery
Although it is interesting enough to find that FAMS
can be used for the validation of genes selected by
other bioinformatic method, it will be better if we can
make use of FAMS for the drug discovery.
3.2.1 Ligand Binding to ”Pocket”
The most popular method to find drug is to find a
small molecule to bind a ”pocket” of each protein. If
FAMS can find or suggest such a candidate for each
of genes in Table 1, it will be very useful.
For example, there are two proteins, MMP8 and
MMP14, in Table 1. They are known to coregulate
target genes(Silva et al., 2012). Both of them are
recognized as members of matrix metalloproteinase
(MMP) family, which is inflammation related protein
family. For MMP8, using 1XUC
A, which is MMP-
13, as a template, FAMS successfully showed that
there are many ligands likely to bind MMP8 (Fig.
2). Similarly, for MMP14, using 1BQO
B, which is
MMP-3, as a template, FAMS successfully showed
that there are many ligands likely to bind MMP14, too
(Fig. 3). Although it is not a finding of a new drug,
this shows the potential for proteins listed in Table 1
which can be new drug targets. Further researches
following this line will be waited.
3.2.2 Termination of Protein Complex
Formation
Other and new possibility of drug target is interrup-
tion in protein complex formation. Many proteins
cannot work as a single substance but can work only
with forming protein complex with other proteins.
Thus, if we interrupt the protein complex formation,
we can also interrupt the function of protein complex.
In Table 2, we have listed protein complex candidates
inferred by FAMS. Since FAMS uses a representative
protein within each cluster having more than 95 % se-
quence similarity as a model protein, there are some-
times more than a thousand model proteins which can
bind to other proteins. We can immediately recognize
that the list includes many reasonable outcomes. For
StructurePredictionwithFAMSforProteinsScreenedCriticallytoAutoimmuneDiseasesbaseduponBioinformatics
265

example, there are 52 model proteins listed between
CSF3 and CSF3R. By name, it is rather obvious that
they are possibly ligand and its receptor. On the other
hand, there are 186 model proteins between CSF3R
and CSF1R. This represents the possibility that each
monomercan form functional protein which can func-
tion together, possibly as a receptor. In addition to
this, both CSF3R and CSF1R most frequently have
non-zero model proteins to bind to each of other refer-
ence proteins. It is reasonable since many can bind to
them as ligand or can form a receptor together. Close
look at this table will give us fruitful information re-
sources to find drug target by the termination of the
formation of protein complex.
In addition to these known and expected protein
complex formation, there are many new findings of
protein complex formation candidates. Fig. 4 shows
one of such possible candidates. In Table 2, there
are 410 possible candidate pairs between CSF1R and
PECAM1. Among these, there is one pair having 61
atom pairs contacting with each other. This means,
there is a structure on PDB (2ZJS) which includes
monomers whose protein structures are expected to be
similar to CSF1R and PECAM1, respectively. 2ZJS
is SecYE translocon, which are expected to func-
tion as a protein-conducting channel(Tsukazaki et al.,
2008). Although this protein complex was found in
Thermus thermophilus, since this kind of proteins are
expected to be highly conserved, it is highly possi-
ble that CSF1R and PECAM1 form protein complex
which is secreted across or integrated into membranes
and play critical role in autoimmune diseases. Thus
if we can find the drug which terminates the protein
complex formation between CSF1R and PECAM1, it
may cure autoimmune diseases.
Predicted protein-protein complex candidates de-
tected are reported in Table 2, but detailed discussion
is deferred due to space constrains. This will be re-
ported in some other opportunity.
4 CONCLUSIONS
In this study, we have demonstrated that how well
FAMS can predict protein structures of candidate
genes which may play critical roles in autoimmune
diseases. Based upon inferred structure, we could an-
notate protein functions, could infer possible ligand
pockets which can bind to proteins, and could find
possible pairs of proteins which can form proten com-
plex, which can be possible candidates of the drug tar-
get. It is confirmed that FAMS can work with other
bioinformatic programs.
REFERENCES
Abd-Allah, S. H., el Shal, A. S., Shalaby, S. M., Pasha,
H. F., Abou el Saoud, A. M., el Najjar, A. R., and
el Shahawy, E. E. (2012). PADI4 polymorphisms
and related haplotype in rheumatoid arthritis patients.
Joint Bone Spine, 79(2):124–128.
Bingle, C. D. and Vyakarnam, A. (2008). Novel innate
immune functions of the whey acidic protein family.
Trends Immunol., 29(9):444–453.
Bouma, B. N., Vlooswijk, R. A., and Griffin, J. H. (1983).
Immunologic studies of human coagulation factor XI
and its complex with high molecular weight kinino-
gen. Blood, 62(5):1123–1131.
Coito, A. J., de Sousa, M., and Kupiec-Weglinski, J. W.
(2000). Fibronectin in immune responses in organ
transplant recipients. Dev. Immunol., 7(2-4):239–248.
Cronstein, B. N. (2007). Interleukin-6–a key mediator of
systemic and local symptoms in rheumatoid arthritis.
Bull NYU Hosp Jt Dis, 65 Suppl 1:S11–15.
Dogliotti, G., Galliera, E., Licastro, F., Porcellini, E., and
Corsi, M. M. (2010). Serum neutrophil gelatinase-
B associated lipocalin (NGAL) levels in Down’s syn-
drome patients. Immun Ageing, 7 Suppl 1:S7.
Fong, L., Ruegg, C. L., Brockstedt, D., Engleman, E. G.,
and Laus, R. (1997). Induction of tissue-specific au-
toimmune prostatitis with prostatic acid phosphatase
immunization: implications for immunotherapy of
prostate cancer. J. Immunol., 159(7):3113–3117.
Frey, T. G., Chan, S. H., and Schatz, G. (1978). Struc-
ture and orientation of cytochrome c oxidase in crys-
talline membranes. Studies by electron microscopy
and by labeling with subunit-specific antibodies. J.
Biol. Chem., 253(12):4389–4395.
Gabr, A. A., Reed, M., Newman, D. R., Pohl, J., Khosla,
J., and Sannes, P. L. (2007). Alterations in cytoskele-
tal and immune function-related proteome profiles in
whole rat lung following intratracheal instillation of
heparin. Respir. Res., 8:36.
Gervin, K., Vigeland, M. D., Mattingsdal, M., Hammer?,
M., Nygard, H., Olsen, A. O., Brandt, I., Harris,
J. R., Undlien, D. E., and Lyle, R. (2012). DNA
methylation and gene expression changes in monozy-
gotic twins discordant for psoriasis: identification
of epigenetically dysregulated genes. PLoS Genet.,
8(1):e1002454.
Heijmans, J., Buller, N. V., Hoff, E., Dihal, A. A., van der
Poll, T., van Zoelen, M. A., Bierhaus, A., Biemond,
I., Hardwick, J. C., Hommes, D. W., Muncan, V., and
van den Brink, G.R. (2012). Rage signalling promotes
intestinal tumourigenesis. Oncogene.
Javierre, B. M., Fernandez, A. F., Richter, J., Al-
Shahrour, F., Martin-Subero, J. I., Rodriguez-Ubreva,
J., Berdasco, M., Fraga, M. F., O’Hanlon, T. P., Rider,
L. G., Jacinto, F. V., Lopez-Longo, F. J., Dopazo,
J., Forn, M., Peinado, M. A., Carreno, L., Sawalha,
A. H., Harley, J. B., Siebert, R., Esteller, M., Miller,
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
266

F. W., and Ballestar, E. (2010). Changes in the pat-
tern of DNA methylation associate with twin discor-
dance in systemic lupus erythematosus. Genome Res.,
20(2):170–179.
Jin, T., Perry, A., Jiang, J., Smith, P., Curry, J. A., Unter-
holzner, L., Jiang, Z., Horvath, G., Rathinam, V. A.,
Johnstone, R. W., Hornung, V., Latz, E., Bowie, A. G.,
Fitzgerald, K. A., and Xiao, T. S. (2012). Struc-
tures of the HIN Domain:DNA Complexes Reveal
Ligand Binding and Activation Mechanisms of the
AIM2 Inflammasome and IFI16 Receptor. Immunity,
36(4):561–571.
Latonen, L., Jarvinen, P. M., Suomela, S., Moore, H. M.,
Saarialho-Kere, U., and Laiho, M. (2010). Ultravi-
olet B radiation regulates cysteine-rich protein 1 in
human keratinocytes. Photodermatol Photoimmunol
Photomed, 26(2):70–77.
Lin, J. X., Li, P., Liu, D., Jin, H. T., He, J., Rasheed, M. A.,
Rochman, Y., Wang, L., Cui, K., Liu, C., Kelsall,
B. L., Ahmed, R., and Leonard, W. J. (2012). Critical
Role of STAT5 Transcription Factor Tetramerization
for Cytokine Responses and Normal Immune Func-
tion. Immunity, 36(4):586–599.
Meylan, E., Burns, K., Hofmann, K., Blancheteau, V., Mar-
tinon, F., Kelliher, M., and Tschopp, J. (2004). RIP1 is
an essential mediator of Toll-like receptor 3-induced
NF-kappa B activation. Nat. Immunol., 5(5):503–507.
O-Sullivan, I., Chopra, A., Carr, J., Kim, T. S., and Co-
hen, E. P. (2008). Immunity to growth factor receptor-
bound protein 10, a signal transduction molecule, in-
hibits the growth of breast cancer in mice. Cancer
Res., 68(7):2463–2470.
O’Hanlon, T. P., Li, Z., Gan, L., Gourley, M. F., Rider,
L. G., and Miller, F. W. (2011a). Plasma proteomic
profiles from disease-discordant monozygotic twins
suggest that molecular pathways are shared in mul-
tiple systemic autoimmune diseases*. Arthritis Res.
Ther., 13(6):R181.
O’Hanlon, T. P., Rider, L. G., Gan, L., Fannin, R., Paules,
R. S., Umbach, D. M., Weinberg, C. R., Shah,
R. R., Mav, D., Gourley, M. F., and Miller, F. W.
(2011b). Gene expression profiles from discordant
monozygotic twins suggest that molecular pathways
are shared among multiple systemic autoimmune dis-
eases. Arthritis Res. Ther., 13(2):R69.
Pan, H. F., Leng, R. X., Tao, J. H., Li, X. P., and Ye, D. Q.
(2011). Ets-1: a new player in the pathogenesis of sys-
temic lupus erythematosus? Lupus, 20(3):227–230.
Quek, A. M., Britton, J. W., McKeon, A., So, E., Lennon,
V. A., Shin, C., Klein, C. J., Watson, R. E., Kotse-
nas, A. L., Lagerlund, T. D., Cascino, G. D., Wor-
rell, G. A., Wirrell, E. C., Nickels, K. C., Aksamit,
A. J., Noe, K. H., and Pittock, S. J. (2012). Autoim-
mune Epilepsy: Clinical Characteristics and Response
to Immunotherapy. Arch Neurol, 69(5):582–593.
Samadani, U. and Costa, R. H. (1996). The transcriptional
activator hepatocyte nuclear factor 6 regulates liver
gene expression. Mol. Cell. Biol., 16(11):6273–6284.
Sarkar, S., Song, Y., Sarkar, S., Kipen, H. M., Laumbach,
R. J., Zhang, J., Strickland, P. A., Gardner, C. R.,
and Schwander, S. (2012). Suppression of the NF-B
pathway by diesel exhaust particles impairs human an-
timycobacterial immunity. J. Immunol., 188(6):2778–
2793.
Selvaraj, R. K., Shanmugasundaram, R., and Klasing, K. C.
(2010). Effects of dietary lutein and PUFA on PPAR
and RXR isomer expression in chickens during an in-
flammatory response. Comp. Biochem. Physiol., Part
A Mol. Integr. Physiol., 157(3):198–203.
Silva, J. A., Ferrucci, D. L., Peroni, L. A., Abrahao, P. G.,
Salamene, A. F., Rossa-Junior, C., Carvalho, H. F.,
and Stach-Machado, D. R. (2012). Sequential IL-23
and IL-17 and increased Mmp8 and Mmp14 expres-
sion characterize the progression of an experimental
model of periodontal disease in type 1 diabetes. J.
Cell. Physiol., 227(6):2441–2450.
Skeie, G. O., Bentsen, P. T., Freiburg, A., Aarli, J. A., and
Gilhus, N. E. (1998). Cell-mediated immune response
against titin in myasthenia gravis: evidence for the in-
volvement of Th1 and Th2 cells. Scand. J. Immunol.,
47(1):76–81.
Taguchi, Y.-h. (2010). A significance test based upon PCA.
SIG-DMSM, A903:38–47.
Tripp, R. A., Barskey, A., Goss, L., and Anderson, L. J.
(2002). Substance P receptor expression on lympho-
cytes is associated with the immune response to res-
piratory syncytial virus infection. J. Neuroimmunol.,
129(1-2):141–153.
Tsukazaki, T., Mori, H., Fukai, S., Ishitani, R., Mori, T.,
Dohmae, N., Perederina, A., Sugita, Y., Vassylyev,
D. G., Ito, K., and Nureki, O. (2008). Conforma-
tional transition of Sec machinery inferred from bac-
terial SecYE structures. Nature, 455(7215):988–991.
Tsunoda, T., Takashima, Y., Tanaka, Y., Fujimoto, T., Doi,
K., Hirose, Y., Koyanagi, M., Yoshida, Y., Okamura,
T., Kuroki, M., Sasazuki, T., and Shirasawa, S. (2010).
Immune-related zinc finger gene ZFAT is an essential
transcriptional regulator for hematopoietic differenti-
ation in blood islands. Proc. Natl. Acad. Sci. U.S.A.,
107(32):14199–14204.
Umeyama, H. and Iwadate, M. (2002). Fams and famsbase
for protein structure. In Current Protocols in Bioin-
formatics. John Wiley & Sons, Inc.
Watson, F. L., Puttmann-Holgado, R., Thomas, F., Lamar,
D. L., Hughes, M., Kondo, M., Rebel, V. I., and
Schmucker, D. (2005). Extensive diversity of Ig-
superfamily proteins in the immune system of insects.
Science, 309(5742):1874–1878.
Zhou, X., Tan, F. K., Xiong, M., Arnett, F. C., and Feghali-
Bostwick, C. A. (2005). Monozygotic twins clinically
discordant for scleroderma show concordance for fi-
broblast gene expression profiles. Arthritis Rheum.,
52(10):3305–3314.
Zoller, K. E., MacNeil, I. A., and Brugge, J. S. (1997).
Protein tyrosine kinases Syk and ZAP-70 display dis-
tinct requirements for Src family kinases in immune
response receptor signal transduction. J. Immunol.,
158(4):1650–1659.
StructurePredictionwithFAMSforProteinsScreenedCriticallytoAutoimmuneDiseasesbaseduponBioinformatics
267
