
Application to Quantify Fetal Lung Branching on Rat Explants
Pedro L. Rodrigues
1,2
, Sara Granja
2
, António Moreira
1,2
, Nuno Rodrigues
3,4
and João L. Vilaça
1,3
1
Algoritmi Center, School of Engineering, University of Minho, Guimarães, Portugal
2
ICVS/3B’s, PT Government Associate Laboratory, Braga/Guimarães, Portugal
3
DIGARC, Polytechnic Institute of Cávado and Ave, Barcelos, Portugal
4
HASLab/INESC TEC, University of Minho, Braga, Portugal
Keywords: Branching Morphogenesis, Lung Development, Image Segmentation, Region-based Algorithms.
Abstract: Recently, regulating mechanisms of branching morphogenesis of fetal lung rat explants have been an
essential tool for molecular research. The development of accurate and reliable segmentation techniques
may be essential to improve research outcomes. This work presents an image processing method to measure
the perimeter and area of lung branches on fetal rat explants. The algorithm starts by reducing the noise
corrupting the image with a pre-processing stage. The outcome is input to a watershed operation that
automatically segments the image into primitive regions. Then, an image pixel is selected within the lung
explant epithelial, allowing a region growing between neighbouring watershed regions. This growing
process is controlled by a statistical distribution of each region. When compared with manual segmentation,
the results show the same tendency for lung development. High similarities were harder to obtain in the last
two days of culture, due to the increased number of peripheral airway buds and complexity of lung
architecture. However, using semiautomatic measurements, the standard deviation was lower and the results
between independent researchers were more coherent.
1 INTRODUCTION
Branching morphogenesis is fundamental to the
growth and development of several organs such as
lung, pancreas, salivary gland, mammary gland, and
kidney and prostate. During the last decades,
analysis of lung branching morphogenesis of fetal
rat explants grown in vitro has been an essential tool
to the research of the underlying molecular and
cellular development mechanisms (Muratore et al.,
2009); (Nogueira-Silva et al., 2008). Therefore, this
methodology has been widely used in many research
centres due to its stability and versatility.
The analysis of branching morphogenesis
involves monitoring lung development in explants
culture, using images acquired at 24-hours intervals
by a stereo microscope during a 5 day period. A
morphometric analysis is usually done to study the
differentiation and growth of lung explants structure.
It quantifies several parameters, such as the inner
and outer epithelial perimeter and area and the
determination of the number of peripheral airway
buds (Nogueira-Silva et al., 2008).
Currently, this analysis is obtained by manual
delineation using generic 2D curves software, which
leads to a time-consuming, dependent on user
expertise and error-prone procedure. Consequently,
this process often results in inaccurate measurements
and forbids the comparison among different
researchers results (Muehlethaler et al., 2008).
Several image processing strategies have been
proposed in the literature to quantify, classify and
segment cellular regions from different image
sources (Yu and Tan, 2009); (Farjam et al., 2007). In
addition, several authors proposed watershed based
algorithms to microscope image processing due to
its ability to produce closed cell contours (Debeir et
al., 2008); (Fan et al., 2008); (Mouelhi et al., 2011).
To best of our knowledge the previous
techniques were never applied to this issue. This
work presents an image processing application that
allows a semiautomatic determination of inner and
outer perimeter of lung braches of rat explants
cultures.
2 METHODS
All methods described below were implemented
under C++ and VTK (Visualization ToolKit).
67
L. Rodrigues P., Granja S., Moreira A., Rodrigues N. and L. Vilaça J..
Application to Quantify Fetal Lung Branching on Rat Explants.
DOI: 10.5220/0004220900670070
In Proceedings of the International Conference on Computer Vision Theory and Applications (VISAPP-2013), pages 67-70
ISBN: 978-989-8565-48-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

2.1 Pre-processing Stage
The aims of this stage are the reduction of the noise
that corrupts the image, while maintaining and
enhancing the relevant object boundaries and
selecting a ROI for further processment.
All lung explant images were acquired with an
Olympus SZX16 stereo microscope in RGB format.
These images were firstly converted to grayscale
values by averaging and normalizing the 3 RGB
components. Then, the grayscale image was input to
an anisotropic diffusion algorithm (implemented as
in (Black et al., 1998)). This algorithm depends on
three parameters, namely the number of iterations (it
= 80), edge parameter (σ=4.5) and a diffusivity
function (Tukey’s biweight as edge-stopping
diffusivity function g(x,σ), Equation 1) (values were
experimentally calculated).
(1)
The anisotropic diffusion algorithm worked as a
Gaussian filter for noise reduction, but with
perseverance of sharper boundaries and image
contours, producing uniformity in the output image
intensity.
Finally, the ROI was selected by removing the
existing noise around the lung explant, by cropping
its region using a labelling and thresholding
algorithm.
2.2 Image Partitioning
Since epithelial boundaries of lung explants are
often characterized by a significant change in image
intensity, an image partitioning yielding meaningful
regions is obtained by detecting all the image edges.
Watershed regions were labelled by a starting
point and follow the flow line, whose direction was
the gradient of intensity, to a local maximum (uphill
direction) or minimum (downhill direction). Both
directions were implemented, but only the downhill
direction was used due to its better performance in
the inner lung explants segmentation.
In the end, the whole image is segmented into
primitive regions where the boundaries of the
watersheds regions coincide with the ridges of the
gradient magnitude surface.
The results of the watershed algorithm are
oversegmented images as illustrated in Figure 1 (A)
and (B).
Figure 1: Result of the watershed partitioning in rat lung
explants.
2.3 Merging Procedure and Inner
Lung Explants Quantification
Although the probability that watershed region
boundaries correspond to boundaries of important
objects increases with oversegmentation, it can also
create many insignificant boundaries. This stage
describes how one dealt with this problem and the
inner lung explants, perimeter and area, were
segmented.
Briefly, this procedure consists on the
identification of regions edges and its connections
with similar intensities, ignoring all others regions
with wide mean intensities variations. It assumes
that:
1. All pixels within the same region are
homogeneous;
2. Regions within lung explant epithelial have
homogeneous intensity variations;
3. Regions within lung explant epithelial are
considerably different from other outside
neighbouring regions;
After establishing the image segmentation through
the application of the watershed algorithm, this
merging procedure performs feature identification in
each region, by identifying the: centroid; mean
intensity distribution (MID); minimum and
maximum values; region edges; edges region
neighbours and the intensity entropy of each region.
Subsequently, the following processing was
considered to produce the final merged and
segmented result:
The user selects one watershed region within the
lung explant epithelial – selected region (SR in red,
Figure 2 (A) and (C));
The boundary information of the SR was
determined in order to found common intersection
edges (IE
i
with i = 1,2,… n, Figure 2 - C) between
neighbourhood regions SRNH;
The MID of each new region (MID_SRNH) was
compared to the MID of the SR (MID_SR). If
MID_SRNH lays between MID_SR±10%, then the
VISAPP2013-InternationalConferenceonComputerVisionTheoryandApplications
68
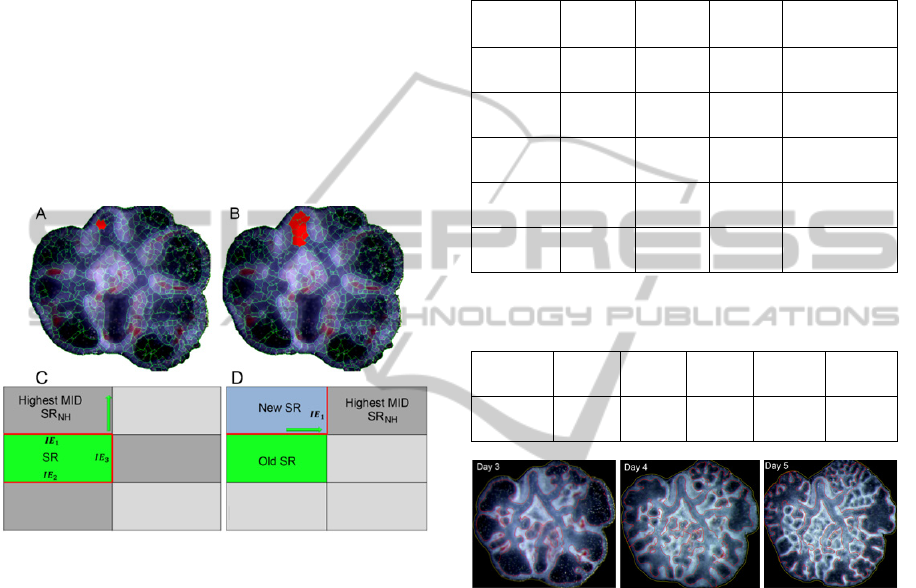
IE
i
(with i = 1,2,… n) is removed and the two
regions are merged; The 10% value was
experimentally calculated by manually evaluating
the mean intensity of the epithelial regions;
If the entropy value of the newly merged region
is not within an interval determined by a 90%
confidence level, which takes into account the pixel
intensity of the two previous regions, these regions
should not have been merged; therefore they are
unmerged and step 2 is repeated in the opposite
direction;
After the merging procedure, the SRNH with the
highest MID similarity value becomes the new SR
and step 2 and 4 are repeated (Figure 2 (B) and (D)).
This algorithm ends when no IE
i
between similar
MID and entropy neighbourhood is found.
Figure 2: Merging procedure overview.
2.4 Outer Lung Explants
Outcome of stage 2.1 was also input to a threshold
algorithm in order to determine the outer lung
explant area and perimeter, by simply counting the
pixels around the object and within it.
3 RESULTS
The suitability and validation of the algorithm was
conducted on stereo microscope images (Olympus
SZX16) acquired at the Life and Health Sciences
Research Institute (ICVS) of School of Health
Sciences, University of Minho -Portugal. These tests
were performed in 50 images, corresponding to
images of 10 sets of lung explants on each day of
culture. All images were previously segmented by
three experienced researchers and this manual
segmentation was used as reference for the
evaluation of the algorithm (some results in Fig. 3).
Each user manually segmented the same image two
times, and the mean and standard deviation of the
inner epithelial perimeter for all sets is shown in
Table 1.
Table 1: Inner epithelial perimeter results obtained by
three different users and by the semiautomatic algorithm.
Culture
Day
User
1
User
2
User
3
Interactive
Method
1
655
± 51
645
± 45
630
± 42
703
± 5
2
841
± 63
850
± 55
830
± 70
954
± 12
3
1565
± 196
1500
± 184
1586
± 177
1752
± 53
4
2202
± 313
2380
± 419
2331
± 450
2592
± 56
5
2482
± 298
2614
± 506
2800
± 357
2887
± 78
Table 2: Summary of the DSC values (%) for each culture
day.
Culture
Day:
1 2 2 4 5
DSC
(%)
92.85
±3.87
91.54
±6.12
90.87
±6.88
88.80
±9.96
86.72
±11.75
Figure 3: Branching morphogenesis in rat lung explants
culture system at each time point. The lines represents the
segmentation results of the inner (red) and outer (yellow)
epithelial perimeter obtained by the automatic algorithm.
The interactive results were also overlapped with the
manual results and its quality and performance were
evaluated using the Dice Similarity Coefficient
(DSC). The mean DSC, of the interactive method
with all the users, from the different days is shown
in Table 2, indicating a successful segmentation
(DSC > 0.85).
4 DISCUSSION
An application for image segmentation was
developed, providing assistance to the researcher
and enabling fast morphometric analysis of lung
explants. The total number of decisions to quantify
ApplicationtoQuantifyFetalLungBranchingonRatExplants
69

morphometric analysis was drastically reduced,
since the user only has to select one single watershed
region.
Generally, best results were obtained in the first
two days of culture with lesser standard deviations.
High similarities between manual and semiautomatic
procedure were harder to get in the last two days of
culture, due to the increased number of peripheral
airway buds and complexity of lung architecture.
The standard deviation of the interactive method was
null after all researchers selected the same start
region to begin the merging process.
The segmentation rate depended on the number
of regions needed to be merged to select the entire
region lung epithelial. However, in all cases the
interactive segmentation time was less than the
manual one (34±7% of the manual time).
The merging procedure was essential to achieve
a good segmentation, since a lot of regions were
firstly created by a watershed algorithm.
Sometimes, the presented method produced
undermerged regions due to ambiguity and lack of
definition of the inner lung explants contours. These
cases increase the probability of merging dissimilar
regions and incoherence between boundaries of
some watershed regions and boundaries of lung
explants inner contours.
5 CONCLUSIONS
Regulating mechanisms of branching morphogenesis
of fetal lung rat explants have been an essential tool
for molecular research. The application of this work
provides a technique for lung rat explants
segmentation and analysis by selecting only one
watershed region belonging to the inner lung
epithelial. The total number of decisions, time-
consumption and user dependence were significantly
decreased.
Further work is needed regarding the merging
procedure and the development of image
enhancement techniques to improve inner lung
epithelial contrast, mainly in the last days of culture,
in order to decrease the standard deviation of results
and increase its reliability. Moreover, a new
algorithm must be developed for counting the
number of peripheral airway buds of lung explants.
ACKNOWLEDGEMENTS
The authors acknowledge to Foundation for Science
and Technology (FCT) - Portugal for the fellowships
with the references: SFRH/BD/74276/2010,
SFRH/BD/68270/2010, SFRH/BPD/46851/2008 and
SFRH/BD/51062/2010.
REFERENCES
Black, M. J., Sapiro, G., Marimont, D. H., Heeger, D.,
1998. Robust anisotropic diffusion. Ieee T Image
Process 7, 421-432.
Debeir, O., Adanja, I., Warzee, N., Van Ham, P.,
Decaestecker, C., 2008. Phase contrast image
segmentation by weak watershed transform assembly.
I S Biomed Imaging, 724-727.
Fan, D., Cao, M. Y., Lv, C. Z., 2008. Wall-adherent cells
segmentation based on cross-entropy and watershed
transform. 2008 International Conference on Audio,
Language and Image Processing, Vols 1 and 2,
Proceedings, 1703-1707.
Farjam, R., Soltanian-Zadeh, H., Jafari-Khouzani, K.,
Zoroofi, R.A., 2007. An image analysis approach for
automatic malignancy determination of prostate
pathological images. Cytometry Part B: Clinical
Cytometry 72B, 227-240.
Mouelhi, A., Sayadi, M., Fnaiech, F., 2011. Automatic
segmentation of clustered breast cancer cells using
watershed and concave vertex graph,
Communications, Computing and Control
Applications (CCCA), 2011 International Conference
on, pp. 1-6.
Muehlethaler V, K. A., Seedorf G, Balasubramaniam V,
Abman SH., 2008. Impaired VEGF and nitric oxide
signaling after nitrofen exposure in rat fetal lung
explants. Am J Physiol Lung Cell Mol Physiol. 294,
110-130.
Muratore CS, L. F., Zhou Y, Harty M, Reichner J, Tracy
TF., 2009. Endotoxin alters early fetal lung
morphogenesis. J Surg Res 155, 225.
Nogueira-Silva C, M. R., Esteves N, Gonzaga S, Correia-
Pinto J., 2008. Intrinsic catch-up growth of
hypoplastic fetal lungs is mediated by interleukin-6.
Pediatr Pulmonol 43, 680-689.
Yu, J., Tan, J., 2009. Object density-based image
segmentation and its applications in biomedical image
analysis. Computer Methods and Programs in
Biomedicine 96, 193-204.
VISAPP2013-InternationalConferenceonComputerVisionTheoryandApplications
70
