
3D Local Binary Pattern for PET Image Classification by SVM
Application to Early Alzheimer Disease Diagnosis
Christophe Montagne, Andreas Kodewitz, Vincent Vigneron, Virgile Giraud and Sylvie Lelandais
University of Evry, IBISC Laboratory, 40 Rue du Pelvoux, CE 1455, 91020 Evry Cedex, France
Keywords:
Local Binary Pattern, Feature Extraction, Positron Emission Tomographic images, Alzheimer disease,
Machine Learning.
Abstract:
The early diagnostic of Alzheimer disease by non-invasive technique becomes a priority to improve the life
of patient and his social environment by an adapted medical follow-up. This is a necessity facing the growing
number of affected persons and the cost to our society caused by dementia. Computer based analysis of
Fluorodeoxyglucose PET scans might become a possibility to make early diagnosis more efficient. Temporal
and parietal lobes are the main location of medical findings. We have clues that in PET images these lobes
contain more information about Alzheimer’s disease. We used a texture operator, the Local Binary Pattern,
to include prior information about the localization of changes in the human brain. We use a Support Vector
machine (SVM) to classify Alzheimer’s disease versus normal control group and to get better classification
rates focusing on parietal and temporal lobes.
1 INTRODUCTION
The number of people affected by the Alzheimer Dis-
ease (AD) is growing. In 2010, this dementia af-
fected 35.6 million people (Wimo and Prince, 2010).
This disease affects patient himself and his social
environment. The detection of the early states of
Alzheimer disease allows to begin some treatments
for the patient and slow down the progress of the dis-
order (Salmon, 2008). Most of works focused on tem-
poral lobes, few of them on parietal lobes (Kodewitz
et al., 2011). Some works on the SPECT-scans (Sin-
gle Photon Emission Computed Tomography) are due
to Ramìrez with a computer-aided diagnosis based
on a selection of image parameters (first and second
order statistics) and Support Vector Machine (SVM)
(Ramìrez et al., 2009). Others methods with PET-
scans (Positron Emission Tomography) are based on
covariance analysis of voxels to classify AD versus
Normal Control (NC) (Scarmeas et al., 2004). The
main idea of this work is that we can automatically
analyse the PET scans to detect AD patient versus
NC patient using textural features. This classifica-
tion can help doctors to make their early diagnostic
and try to localize some brain areas where Alzheimer
Disease is growing. In our study, we use 3D-scans
extracted from the ADNI database (Alzheimer’s Dis-
ease Neuroimaging Initiative) and we propose to use
a textural operator which is the Local Binary Pattern
(LBP) (Pietikäinen and Ojala, 2000) extended to the
3D. Then this new feature was studied by an ANaly-
sis Of VAriance (ANOVA). The analysis highlighted
a link between LBP patterns and the type of patient
or some brain areas. Based on this new feature, we
train a machine learning algorithm SVM to classify
AD versus NC PET-scans. For this step, in a first time
we use as Region Of Interest (ROI) parietal and tem-
poral lobes, and then, in a second time full PET-scans
are considered.
In the first section we describe the materials used
in this study. Secondly we introduce the 3D Local
Binary Pattern that we developed. Then we explain
the ANOVA method and the obtained results. Finally
after a short presentation of SVM, we discuss on the
results of the classification using the LBP caracteris-
tics and we conclude.
2 MATERIALS
In this study, PET-scan provides three-dimensional
functional imaging data that measures the metabolism
in the brain which are acquired by a non-invasive
method, Figure 1 shows how the AD reduces the
metabolism in the brain. This scan is obtained by 18F-
145
Montagne C., Kodewitz A., Vigneron V., Giraud V. and Lelandais S..
3D Local Binary Pattern for PET Image Classification by SVM - Application to Early Alzheimer Disease Diagnosis.
DOI: 10.5220/0004226201450150
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2013), pages 145-150
ISBN: 978-989-8565-36-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
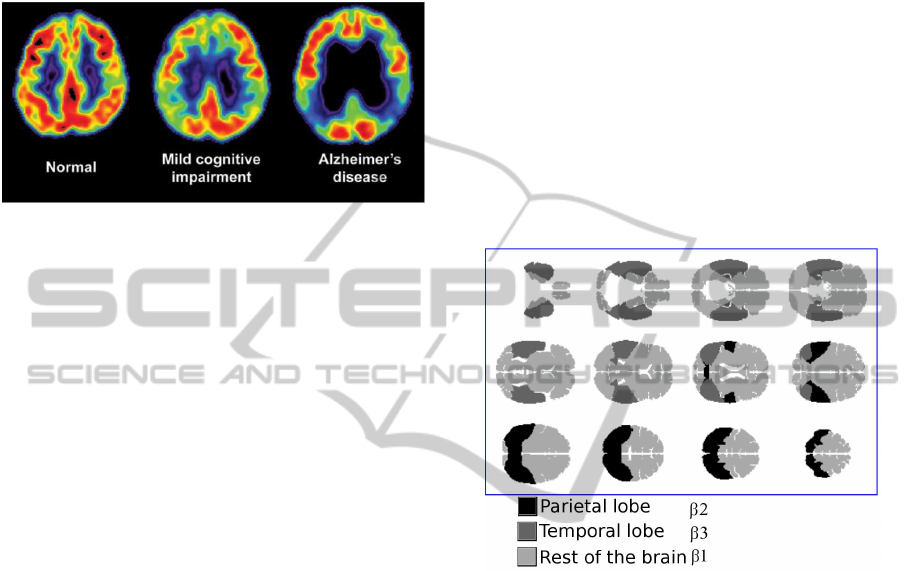
fluorodesoxyglucose (in short 18F-FDG) injection to
a patient. This radioactive isotope allows to capture
the distribution of brain activity. With this technique,
we can detect the first abnormalities before structural
alteration (Bennys et al., 2001).
Figure 1: PET scans of AD, MCI and NC patient.
2.1 ADNI Database
The American Alzheimer’s Disease Neuroimaging
Initiative (ADNI, http:// www.loni.ucla.edu/ ADNI/)
collects data of patients affected by AD, mild cog-
nitive impairment (MCI) and normal control group
(NC). This includes diagnosis based on mental state
exams (e.g. mini mental state exam (MMSE)),
biomarkers, MRI and PET scans. In this database we
chose PET scans with 18F-FDG tracer from AD and
NC patients. Each image contains 91×109 ×91 vox-
els after normalization (the PET-scans have initially
different sizes). For our purpose we extracted 166 3D-
scans (one 3D-scan for one patient) which are sorted
in 82 AD and 84 NC. Moreover, the AD scans are
taken early in the disease to simulate a situation of
early diagnosis.
2.2 Brain Atlas
Today, the definite diagnosis of AD is based on the
post-mortem observation of intracellular neurofibril-
lary tangles (NTF): β-amyloid deposition in the form
of extracellular senile plaques and blood vessel de-
posits, synapse dysfunction and loss. NTF deposition
originates in the medial temporal lobes and then be-
gins to cluster in the adjacent inferior temporal and
posterior cingulate cortex in mild AD, and finally
spreads to the parieto-temporal and prefrontal associ-
ation cortices. Medical doctors use this information in
their analysis of PET scans. They search for abnormal
variations in the brain metabolism and do their diag-
nosis based on the location of these abnormalities. In
order to emulate this method, we decide to use a brain
atlas which gives us a brain model. With the
c
Matlab
toolbox WFU PickAtlas (Maldjian et al., 2003), using
the Talairach daemon by Lancaster et al. (Lancaster
et al., 2000), it is even possible for nonspecialists to
select a volume of interest in the brain by knowing
the name of a certain area, e.g. the name of the lobe,
and create an indexed mask for this volume of inter-
est. We used these prerequisites to create a simplified
map which distinguishes only three zones: temporal
lobes, parietal lobes and the rest of the brain (see fig-
ure 2). Each voxel of a PET-scan matches with a voxel
of this atlas because our atlas size is the same as our
normalised brain PET-scans. So we have a unique
model of brain for all individuals. Parietal and tem-
poral areas are considered identical from one brain
to another and adopt a location and volume average.
Parietal lobes contain 26850 voxels, temporal lobes
contains 33103 voxels and rest of the brain contain
119907 voxels in each PET-scan.
Figure 2: Brain atlas.
3 LOCAL BINARY PATTERN
3.1 2D Local Binary Pattern
LBP was proposed by Pietikäinen et Ojala in the 90’s
(Pietikäinen and Ojala, 2000). The idea is to give a
pattern code to each pixel. Gray level of the central
pixel g
c
is compared to gray level of each neighboring
pixel g
i
. Then by thresholding, a value of 0 or 1 is
associated to each neighboring pixel.
LBP
2d
P,R
=
P−1
∑
i=0
s(g
i
− g
c
)2
i
, with s(x) =
(
1 if x ≥ 0
0 else.
(1)
where P is the number of neighbor pixels separated
from R pixel(s) to the central pixel c. Figure 3 illus-
trates this feature.
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
146
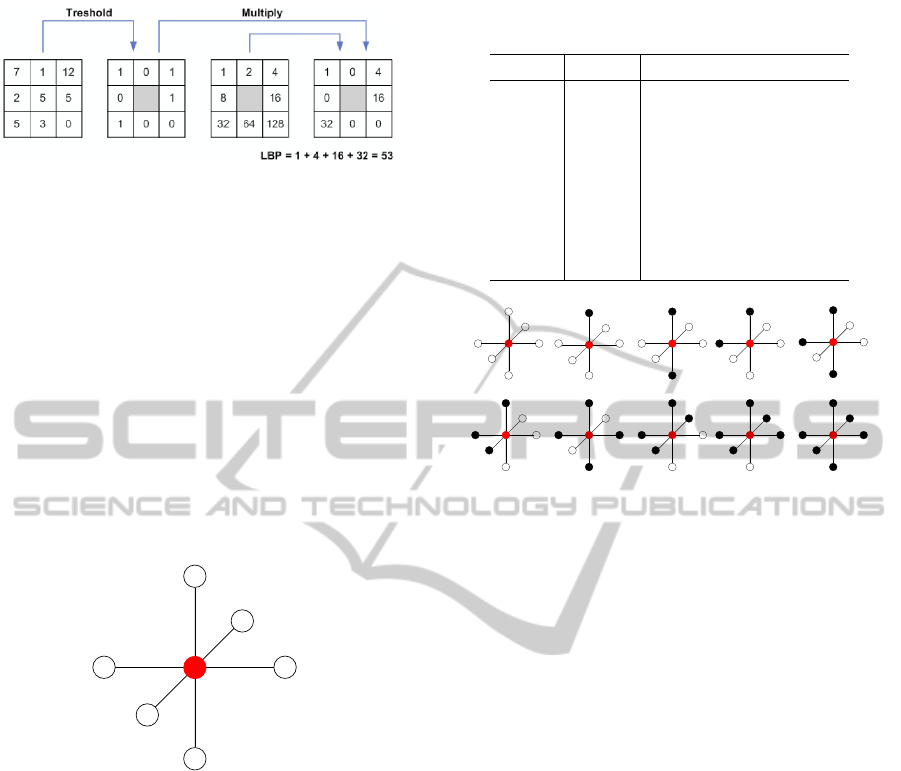
Figure 3: Example of 2D-LBP with P = 8 and R = 1.
3.2 3D Local Binary Pattern
The main idea of this work is an adaptation of the LBP
to capture the 3D structure of texture. We need LBP
patterns which have a rotation invariance and qualify
the AD PET-scans. Some works have been performed
on the extend of the Local Binary Pattern method for
characterisation of 3D textures by L. Paulhac (Paul-
hac et al., 2008). His vision of 3D-LBP consists on
a superposition of circles that represent a spherical
neighborhood. Each circle can be encoded like a 2D-
LBP. Our proposition consists to select 6 nearest vox-
els and to order them for encoding pattern (see figure
4).
g0
g1
g2
g3
g4
g5
Figure 4: 3D-LBP with 6 neighbors LBP
3d
6,1
.
By this way, we have 2
6
= 64 possible patterns
that we merge in 10 groups according to geometrical
similarities (see figure 5). Each group is filled with
patterns which have the same numbers of neighbor
voxels with a gray level higher than the central voxel
c. The idea was to keep a rotation invariance in each
group. Table 1 defines these groups with:
card(c) =
P−1
∑
i=0
s(g
i
− g
c
) (2)
where P = 6 is the number of voxels in neighborhood
and R = 1 or R = 2 the distance between the central
voxel and its neighbors. We use two values of R to
capture micro and macro-structure of the texture with
our 3D-LBP. In equation 2, card(c) gives the number
of neighbors with an higher gray level than the central
voxel.
Table 1: Definition of the 10 groups of patterns rotationally
invariant.
LBP
3d
P,R
card(c) condition
1 0
2 1
3 2 opposite voxels
4 2 bend voxels
5 3 voxels on the same plane
6 3 voxels on different planes
7 4 voxels on the same plane
8 4 voxels on different planes
9 5
10 6
1(1) 3(3)2(6)
9(6)8(12) 10(1)
6(8)
7(3)
4(12) 5(12)
Figure 5: Merging of 2
6
= 64 patterns in 10 groups. The
integer in brackets indicates the number of different patterns
for each group.
In our experimental protocol, the count of these
patterns in ROI of PET-scans or in full PET-scans will
be the input variable of our machine learning algo-
rithm.
3.3 ANOVA
The 3D-LBP features may be an important character
for the classification of Alzheimer PET-scan. We per-
form an analysis of variance to find the link between
a variable of interest (the count of 3D-LBP features)
and some response variables (type of patient, brain
area, type of pattern) which are also called factors and
its relevance.
Suppose that the variable to explain Y follows a
normal distribution in each population. y
i j
was a re-
alization of Y ∼ N (µ
i
,σ). There are two hypothe-
sis: H
0
-Null hypothesis, every data follows a normal
distribution; H
1
-At least one sample follows an other
normal distribution.
There are 3 factors:
• α
u
: type of patient (AD/NC),
• β
v
: brain area (temporal lobe β
3
, parietal lobe β
2
,
rest of brain β
1
, full brain β
0
) (see figure 2),
• γ
w
: type of pattern (1 .. . 10) (see figure 5).
We search the influence of a factor set (α
u
, β
v
, γ
w
)
on the variable of interest Y . This method can be ap-
3DLocalBinaryPatternforPETImageClassificationbySVM-ApplicationtoEarlyAlzheimerDiseaseDiagnosis
147

plied on each factor or on all together. For example,
Eqs. 3 and 4 show a case with a single factor.
y
ik
= µ + m
α
i
+ ε
ik
(3)
where µ is an offset, m
α
i
the theoretical mean of
the sample i and ε
ik
are Gaussian i.i.d error terms
(N (0,σ
2
)).
y
11
.
.
.
y
1n
1
y
21
.
.
.
y
2n
2
.
.
.
=
1 0 0
.
.
.
.
.
.
.
.
.
1 0 0
0 1 0
.
.
.
.
.
.
.
.
.
0 1 0
.
.
.
.
.
.
.
.
.
m
α
1
m
α
2
m
α
3
+
ε
11
.
.
.
ε
1n
1
ε
21
.
.
.
ε
2n
2
.
.
.
(4)
where m is the theoretical mean of the sample. This
equation shows the variance decomposition:
3
∑
i=1
n
i
∑
k=1
(X
ik
− m)
2
=
3
∑
i=1
n
i
(m
i
− m)
2
+
3
∑
i=1
n
i
∑
k=1
ε
2
ik
(5)
SC
2
T
= SC
2
A
+ S
2
R
m
α
i
and m are estimated by: ¯y
i
=
∑
3
i=1
y
ik
and ¯y =
∑
3
i=1
∑
n
i
k=1
y
ik
. Under H
0
, we have:
SC
T
σ
2
∼ χ
2
n−1
,
SC
A
σ
2
∼ χ
2
3−1
,
S
R
σ
2
∼ χ
2
n−3
(6)
and:
T =
SC
2
A
/(3 − 1)
S
2
R
/(n − 3)
∼ F
p−1,n−p
. (7)
Under H
0
, the statistics T follows a Fisher distri-
bution with (3 − 1,n − 3) degrees of freedom. The
critical value is T > F
p−1,n−p,1−α
, where α settles the
confidence level with which one can reject the hy-
pothesis H
0
(by convention often chosen to be 95%).
Table 2 exposes a part of the results. The p-value
of each group of pattern on each area of brain is dis-
played.
The p-value is the probability of obtaining a test
statistic at least as extreme as the one that was actu-
ally observed, assuming that the H
0
hypothesis is true.
The lower is the p-value, the higher is the probabil-
ity of H
1
to be true. We choose a p-value threshold
to 0.005: we accept H
1
is true with 99.5% or more
(bold p-values in the table). We can see p-values cor-
responding to the full brain (β
0
) or rest of the brain
(β
1
) are higher than our threshold: the count of the
3D-LBP patterns is not informative in these areas. In
the parietal and temporal lobes (β
2
, β
3
), we notice the
Table 2: p-values for the different brain areas.
LBP Group
number
Pr(> F)
β
0
β
1
β
2
β
3
1 2.13e-1 1.42e-1 5.83e-5 2.63e-2
2 5.47e-1 3.14e-1 1.66e-5 1.86e-4
3 2.71e-1 6.38e-1 3.14e-4 2.51e-7
4 7.89e-1 1.60e-1 1.76e-3 1.07e-2
5 9.77e-1 2.24e-1 8.60e-5 4.69e-3
6 7.61e-1 4.20e-1 1.56e-3 5.72e-3
7 9.22e-1 3.38e-1 7.40e-3 1.39e-1
8 7.62e-1 4.13e-1 2.50e-3 1.79e-3
9 6.00e-1 2.12e-2 3.22e-3 3.69e-1
10 3.49e-1 2.54e-2 1.86e-1 6.43e-2
power of some LBP patterns for discriminating the
type of patient α
i
.
These observations allow to use machine learning
to create a classifier with the 3D LBP features ex-
tracted from the PET-scan.
4 CLASSIFICATION BY
SUPPORT VECTORS MACHINE
The SVM was introduced by Vapnik (Vapnik, 1998).
We used a description of the SVM which is based on
Schölkopf and Smola (Schölkopf and Smola, 2002)
works. Consider a training data set consisting of two
separable classes in a n-dimensional feature space.
This means each class forms its own cluster and those
clusters do not intersect. From this follows that there
exists at least one hyperplane able to separate the two
classes with one class on each side of the hyperplane.
We use a Gaussian kernel in SVM, γ ≥
0, k(x, x
0
) = e
(−||x−x
0
||
2
/γ
2
)
. In figure 6 we can see
the contour plot of the error landscape resulting from
a grid search on hyperparameter cost C and gamma γ.
C is a cost constant for the Lagrangian. This grid is
obtained with the function tune in R. The red circle
shows the area with the best parameters.
PET-scans used in this study come from the ADNI
database. Each scan contains 91 × 109 × 91 voxels.
We selected 82 patients from AD group and 84 from
NC group (one patient = one PET-scan). We try to
classify AD versus NC. Each voxel in the image is
encoded by LBP
3d
6,1
and LBP
3d
6,2
. We have 10, 20 or
30 input variables if we use 1, 2 or 3 areas of brain
(parietal β
2
, temporal β
3
, rest of the brain β
1
) in the
classifier. With full brain (β
0
) we have 10 variables.
And when we use the LBP
3d
6,1
and LBP
3d
6,2
, the number
of variables is doubled.
These variables constitute our input vector for
SVM. In table 3 we see the classification rate for the
3 brain areas or for parietal and temporal lobes. Other
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
148
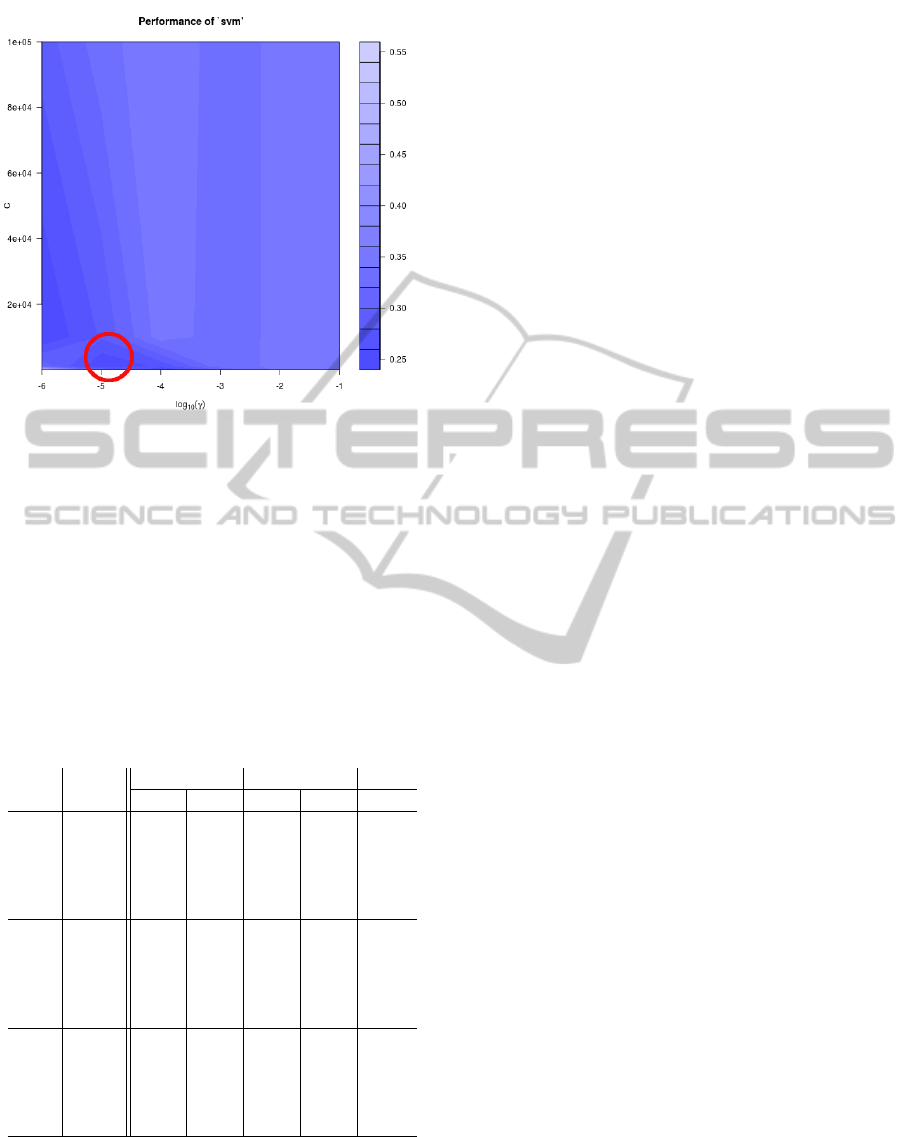
Figure 6: Visualisation of the tuning parameters C and γ.
results with only one brain area (β
1
, β
2
or β
3
) or full
brain (β
0
) don’t appear in this table because of lower
results. The cross validation consists in a number of
runs equal to size of the base of a SVM. For each run
we divide the main database with a part used for train-
ing and the other for testing. The mean of all these
runs give us the rate of cross validation.
Table 3: Precision, recall and cross validation (CV) for
SVM classification - ¬: LBP
3d
6,1
, : LBP
3d
6,2
. β
1,2,3
is put
for the use of all brain areas; β
2,3
is for temporal and pari-
etal lobes.
LBP Brain Precision Recall
type areas AD NC AD NC CV
β
1
68% 70% 70% 68% 60%
β
2
67% 70% 70% 67% 68%
¬ β
3
75% 71% 67% 79% 64%
β
1,2,3
89% 81% 79% 90% 75%
β
2,3
85% 79% 76% 86% 66%
β
1
64% 67% 68% 63% 56%
β
2
73% 70% 66% 76% 70%
β
2
65% 71% 68% 76% 66%
β
1,2,3
87% 79% 76% 89% 75%
β
2,3
82% 82% 81% 83% 68%
β
1
74% 77% 77% 74% 63%
¬ β
2
74% 70% 67% 77% 69%
& β
3
77% 72% 67% 81% 67%
β
1,2,3
87% 79% 76% 89% 77%
β
2,3
82% 82% 81% 83% 69%
Table 3 shows results obtained with different vari-
ables of LBP
3d
6,1
and LBP
3d
6,2
. We can notice that all the
results are very close together. But when we use the
3 brain areas, the SVM classifier reaches better rates
than just using parietal and temporal lobes. The best
CV rate is 77% with LBP
3d
6,1
&LBP
3d
6,2
. We can assume
that this value is relatively low because the scans cor-
respond to the beginning of the disease when the di-
agnosis is more difficult than for a patient severely
affected.
5 CONCLUSIONS
We propose a new method to identify patients af-
fected by the Alzheimer Disease from their PET-scan.
With the ANOVA we have shown LBP are impor-
tant features to class AD versus NC PET-scans. Then
we train and test a classifier to create an automatic
computer-aided diagnosis for the Alzheimer Disease
using the LBP caracteristics extracted from the PET-
scan. We reach to a score of 77% by using SVM
method with LBP
3d
6,1
and LBP
3d
6,2
patterns.
Currently we improve this work by slicing in
small cubes, in each of these block we count the LBP
patterns. Then these counts are used as variables for a
Random Forest classification. RF is adapted to com-
pute and to have a good classification rate with this
high number of features.
An other way to improve the results can be to de-
velop some new LBP types to capture more accurately
the texture of brain in the PET-scans. We can notice
that some regions that are outside temporal and pari-
etal lobe contain also a certain amount of discrim-
inating information which is captured with our ap-
proach. In this case we can extend the analysed brain
areas to find other ROI. A brain mapping of each PET-
scan can be used to define more cleverly position of
the temporal and parietal lobes, or some other sub-
parts. The idea is to customize an atlas for each pa-
tient rather than our standard atlas for all the database.
This way is already used on IRM-scans. With this, we
have to segment brain to find the edges of the differ-
ent areas. In the same way, merge IRM and PET data
will be an interesting search.
ACKNOWLEDGEMENTS
Data used in the preparation of this arti-
cle were obtained from the Alzheimer’s Dis-
ease Neuroimaging Initiative (ADNI) database
(www.loni.ucla.edu/ADNI). As such, the investi-
gators within the ADNI contributed to the design
and implementation of ADNI and/or provided data
but did not participate in analysis or writing of
this report. ADNI investigators include (complete
listing available at http://www.loni.ucla.edu/ADNI/
Collaboration/ADNI_Manuscript_Citations.pdf).
3DLocalBinaryPatternforPETImageClassificationbySVM-ApplicationtoEarlyAlzheimerDiseaseDiagnosis
149

REFERENCES
Bennys, K., Rondouin, G., Vergnes, C., and Touchon,
J. (2001). Diagnostic value of quantitative EEG
in alzheimer’s disease. Clinical Neurophysiology,
31(3):153–160.
Kodewitz, A., V., V., Montagne, C., and Lelandais, S.
(2011). Where to search for alzheimer’s disease re-
lated changes in pet scans? RITS, Rennes, France.
Lancaster, L., Woldorff, M. G., and Parsons, L. M. (2000).
Automated talairach atlas labels for functional brain
mapping. Human Brain Mapping, 10:120–131.
Maldjian, J. A., Laurienti, P. J., Burdette, J. H., and
Kraft, R. A. (2003). An automated method for neu-
roanatomic and cytoarchitectonic atlas-based interro-
gation of fmri data sets. NeuroImage, 19:1233–1239.
Paulhac, L., Makris, P., and Ramel, J. (2008). Compari-
son between 2d and 3d local binary pattern methods
for characterisation of three-dimensional textures. In
Series, B., editor, Lecture Notes in Computer Science,
volume 5112/2008, pages 670–679, Pòvoa de Varzim,
Portugal. ICIAR.
Pietikäinen, M. and Ojala, T. (2000). rotation-invariant tex-
ture classification using feature distributions. Pattern
Recognition, 33:43–52.
Ramìrez, J., Gòrriz, J., and et al., D. S.-G. (2009).
Computer-aided diagnosis of alzheimer’s type demen-
tia combining support vector machines and discrimi-
nant set of features. Information Sciences.
Salmon, E. (2008). Différentes facettes de la maladie de
type azheimer. Rev Med Liege, 63(5-6):299–302.
Scarmeas, N., Habeck, C. G., and et al., E. Z. (2004). Co-
variance pet patterns in early alzheimer’s disease and
subjects with cognitive impairment but no dementia:
utility in group discrimination and correlations with
functional performance. NeuroImage, 23(1):35 – 45.
Schölkopf, B. and Smola, A. (2002). Learning with Ker-
nels - Support Vector Machines, Regularization, and
Beyond. The MIT Press.
Vapnik, V. (1998). Statistical learning theory. Wiley.
Wimo, A. and Prince, M. (2010). World alzheimer report
2010. http://www.alz.co.uk/research/world-report.
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
150
