
A Signal-independent Algorithm for Information Extraction
and Signal Annotation of Long-term Records
Rodolfo Abreu
1
, Joana Sousa
2
and Hugo Gamboa
1,2
1
CEFITEC, Departamento de F´ısica, FCT, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal
2
PLUX - Wireless Biosignals S.A., Lisbon, Portugal
Keywords:
Biosignals, Waves, Events Detection, Features Extraction, Pattern Recognition, k-Means, Parallel Computing,
Signal Processing.
Abstract:
One of the biggest challenges when analysing data is to extract information from it. In this study, we present a
signal-independent algorithm that detects events on biosignals and extracts information from them by applying
a new parallel version of the k-means clustering algorithm. Events can be found using a peaks detection algo-
rithm that uses the signal RMS as an adaptive threshold or by morphological analysis through the computation
of the signal meanwave. Different types of signals were acquired and annotated by the presented algorithm.
By visual inspection, we obtained an accuracy of 97.7% and 97.5% using the L
1
and L
2
Minkowski distances,
respectively, as distance functions and 97.6% using the meanwave distance. The fact that this algorithm can
be applied to long-term raw biosignals and without requiring any prior information about them makes it an
important contribution in biosignals information extraction and annotation.
1 INTRODUCTION
The main goal of clustering is to extract features from
data objects that will allow data to be divided into
clusters where objects in the same cluster have a max-
imum homogeneity (Hansen and Jaumard, 1997).
Applying clustering techniques to biosignals is an
approach that has been used recently. Clustering on
electrocardiography (ECG) signals has been used to
group the QRS complexes (or beats) into clusters that
represent central features of the data (Cuesta-Frau
et al., 2002). Also in electromyography (EMG), clus-
tering algorithms have been used to cluster data fea-
tures which will be used as input of a classifier, allow-
ing a high training speed (Chan et al., 2000).
The developed algorithm aims at extracting infor-
mation from biosignals and annotate them by apply-
ing clustering techniques. For that, the detection of
signal events is required. In order to accomplish this,
a peak detection algorithm that thresholds above the
signal root mean square (RMS) level and the com-
putation of the signal meanwave by calculating the
mean value for each time-sample of the signal cycles
(Nunes et al., 2011) were used.
Then, our algorithm takes distance measures us-
ing different distance functions that will be used as
input for a new parallel version of the k-means clus-
tering algorithm. The main concept of the k-means al-
gorithm was kept, which is a partitioning method for
clustering where data is divided into k partitions (War-
ren Liao, 2005). The optimal partition of the data is
obtained by minimizing the sum-of-squared error cri-
terion with an interactiveoptimization procedure. Our
clustering algorithm divides the observations to be
clustered into parts, performs k-means for each part
and finally assembles the results. Thus, in this paper
we present an approach that allows long-term signal
classification without any prior information and with
fast speed performance due to the employment of par-
allel computing techniques.
2 SIGNAL PROCESSING
ALGORITHMS
2.1 Data Acquisition
For the acquired biosignals, a triaxial accelerometer
sensor (xyzPLUX), an ECG sensor (ecgPlux), a BVP
sensor (bvpPlux), and EMG sensor (emgPlux) and a
respiratory sensor (respPlux) were used. These sen-
sors were connected to a device – bioPlux research
unit – responsible for the signal analog-to-digital con-
323
Abreu R., Sousa J. and Gamboa H..
A Signal-independent Algorithm for Information Extraction and Signal Annotation of Long-term Records.
DOI: 10.5220/0004241303230326
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2013), pages 323-326
ISBN: 978-989-8565-36-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

version and bluetooth transmission to the computer.
Signals were sampled at a 1000 Hz frequency and
converted using a 12 bit ADC (PLUX, 2012).
The ECG signals were obtained in different con-
texts. A 7 hour signal was acquired during a night of
sleep of a person diagnosed with amyotrophic lateral
sclerosis under the project wiCardioResp. One ECG
was also acquired under the project ICT4Depression
where patients with depression are monitored at
home. An ACC signal where the subject was walking
at average speed was also acquired under this project.
For research and evaluation purposes, one respiratory
signal was acquired. Besides, one BVP signal was
acquired right after a subject performed some exer-
cise and then, at rest. Finally, two scenarios (Act1:
Walk, Run, Walk, Jump; Act2: Crouching, leg flex-
ion, leg elevation) were created enabling the acquisi-
tion of ACC signals with different modes. Both activ-
ities were performed by a single subject. From Activ-
ity 2, an EMG signal was also acquired.
2.2 Algorithm Implementation
2.2.1 Events Detection
As it was stated in the previous section, the first step
in our algorithm is to detect events in cyclic biosig-
nals. We propose two different methods for events
detections which will be described next.
Peaks Detection Approach. In our approach, we
define the threshold as being the RMS of the signal.
In order to obtain a higher accuracyin detecting signal
events, our algorithm updates the threshold every ten
seconds. Due to its simplicity and low computational
cost, using the signal RMS as an adaptive threshold
for peaks detections is an interesting method for ac-
complishing the first step of our algorithm.
Meanwave Approach. In this approach, the basic
concept was previouslyimplemented by (Nunes et al.,
2011). The main goal of the autoMeanWave algo-
rithm is to separate the cycles from a periodic biosig-
nal. For that, the cycles (or waves) size – winsize – is
estimated by computing the fundamental frequency,
f
0
, of the signal. Then, the events are detected and the
meanwave is computed. The signal events are aligned
using a notable point from the meanwave.
The main concept of this algorithm was kept but
some improvements were made. In fact, a time-
domain method for f
0
estimation based on the auto-
correlation of finite time series was used. The events
alignment step was also improved by adding a second
phase of alignment which is wave-specific. In fact,
after performing the alignment by choosing a notable
from the meanwave, our algorithm runs through all
the signal waves and relocates the events to the same
notable point from each wave. Finally, the ability to
process long-term biosignals was achieved by divid-
ing the signal into parts and detecting the events in
each part individually. To guarantee that no informa-
tion was lost among transition zones, a f
0
-dependent
overlap was introduced.
2.2.2 Distance Functions and Distance Measures
In order to obtain inputs to the parallel k-means algo-
rithm, a set of different distance functions was used.
First of all, the Minkowski-form Distance defined as
(Chan et al., 2000)
L
p
(P,Q) =
∑
i
|P
i
− Q
i
|
p
!
1/p
, 1 ≤ p ≤ ∞ (1)
In this study, the L
1
, L
2
and L
∞
distance functions
were used and the squared version of L
2
, L
2
2
, also. Be-
sides, the χ
2
histogram distance was also tested.
In order to obtain distance measures that will be
used as inputs for a clustering algorithm, the com-
puting of a distance matrix it is usually necessary.
This distance matrix is obtained by computing the dis-
tance between each observation and all the other ones.
However, the order relationship between two consec-
utive samples, which is a property of time series, al-
lows morphological comparisons between waves (or
cycles of a signal), w
i
, by simply computing a dis-
tance array where each element, d
i
, is given by:
d
i
= f(w
i
,w
i+1
), i = 1,... , n − 1 (2)
being f the distance function and n the number of
waves. w
i
can also represent the meanwave but, in
this case, i = 1,...,n.
Although the distance matrix carries richer infor-
mation about waves resemblance than the distance ar-
ray, its high computational cost makes it impossible
to be used in long records.
2.2.3 Clustering Algorithm
In our algorithm, the observations to be clustered
are divided into N parts. Then, the k-means algo-
rithm is applied in each part and a set of centroids
[a
i
,b
i
,...,k
i
] are computed, with i = 1, . . . , N being
number of each part and k the number of partitions
given as input for the k-means algorithm. Since the
k-means algorithm randomly assigns clusters to the
computed k partitions, different clusters assignment
is obtained. By assembling all the N sets of cen-
troids, a new set of observations is computed. By run-
ning one last time the k-means algorithm, the global
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
324
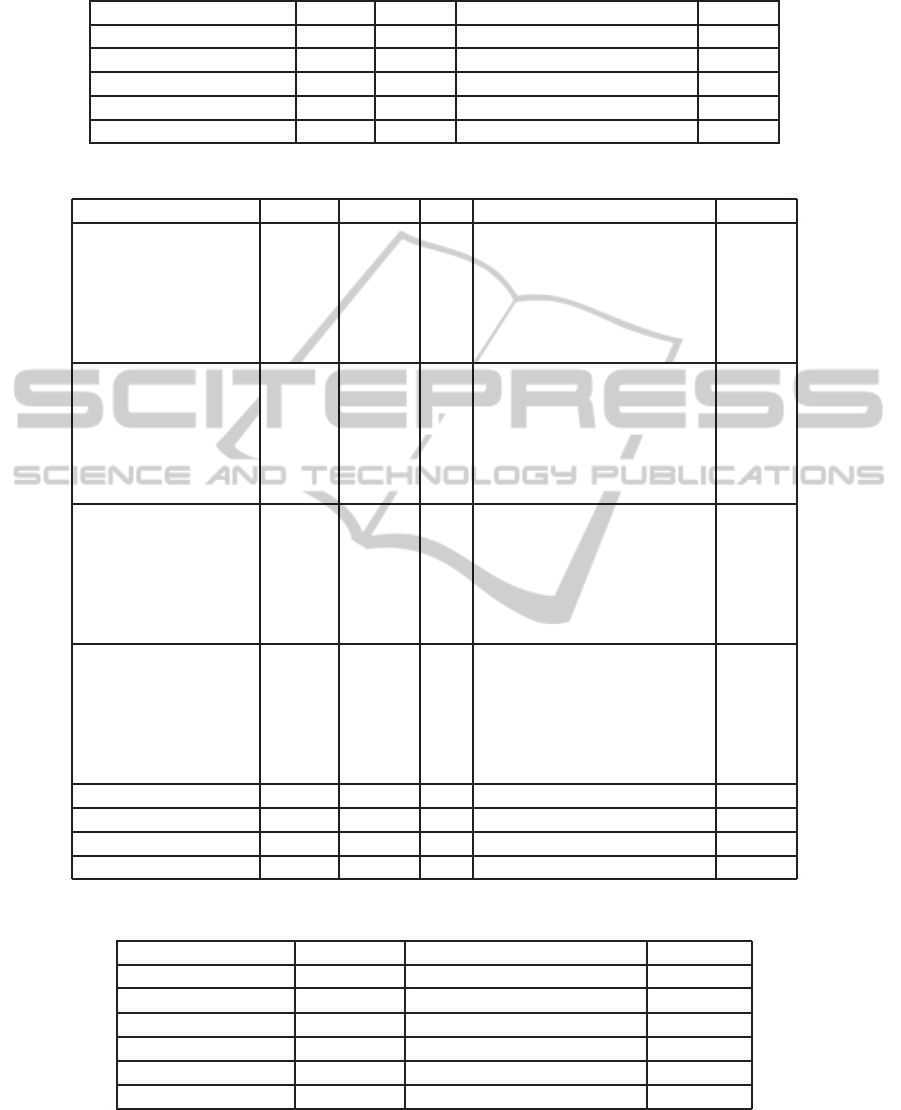
Table 1: Clustering results using ∆t
i
with k = 2 clusters.
Signal Cycles Misses Correctly clustered cycles Errors
ECG
1
(wiCardioResp) 24551 24279 0 272
ECG
2
(Walking - ICT) 199 198 0 1
BVP (Rest/Exercise) 165 162 2 1
ACC
1
(Walking - ICT) 132 131 0 1
Respiration 67 65 1 1
Table 2: Clustering results using morphological comparison with k clusters.
Signal Cycles Misses Correctly clustered cycles Errors
ECG
1
(k = 2) 24551 272
L
1
24028 251
L
2
2
23579 700
L
2
23998 281
L
∞
23447 832
χ
2
23565 714
Mw 24021 258
ECG
2
(k = 2) 199 1
L
1
191 7
L
2
2
178 20
L
2
193 5
L
∞
172 26
χ
2
182 16
Mw 193 5
BVP (k = 2) 165 1
L
1
123 41
L
2
2
98 66
L
2
124 42
L
∞
111 53
χ
2
94 70
Mw 114 50
Respiration (k = 2) 67 1
L
1
65 3
L
2
2
64 4
L
2
46 19
L
∞
44 21
χ
2
60 5
Mw 52 13
ACC
1
(k = 2) 185 1 Mw 179 5
ACC
2
(Act 1, k = 2) 672 1 Mw 666 5
ACC
3
(Act 2, k = 3) 56 1 Mw 53 2
EMG (Act 2, k = 3) 56 1 Mw 47 8
Table 3: Accuracy obtained using different distance functions for the clustering algorithm’s input.
Distance Function All Cycles Correctly clustered cycles Accuracy
L
1
24982 24407 97.7%
L
2
2
24982 23919 95.7%
L
2
24982 24361 97.5%
L
∞
24982 23774 95.2%
χ
2
24982 23901 95.7%
Mw 25951 25325 97.6%
centroids that represent the data as a whole are com-
puted. Finally, the Euclidean Distance is computed
between each centroid and each observation, result-
ing in a k×M matrix. Searching for the line where the
minimum element of each row is located, the cluster
which that observation will be assigned to is provided.
ASignal-independentAlgorithmforInformationExtractionandSignalAnnotationofLong-termRecords
325

3 RESULTS AND DISCUSSION
A visual inspection for performance evaluation was
taken and different criteria were used for the differ-
ent types of clustering results. The concepts of error
(when a cycle is wrongly identified or classified) and
miss (when a cycle is not classified) are used in both
types of results. Only the meanwave approach was
used to obtain the signal events.
3.1 Clustering using Time-samples
Difference Information
Despite its conceptual simplicity, an almost perfect
events detection and alignment can lead to a time-
samples variability analysis between those events. In
fact, this information is useful if the main goal is to
separate parts of the signal where significant changes
in frequency are observed. After running this cluster-
ing method in order to divide the data into k partitions,
the obtained results are presented in Table 1.
For the ECGs, a heart rate variability (HRV) anal-
ysis should be taken to assess the clustering results.
3.2 Clustering using Morphological
Comparison
Next, a morphological analysis was taken in order to
obtain signal annotations. Table 2 shows the obtained
results for each signal and Table 3 accounts for the ob-
tained accuracy using the various distance functions.
In ECG
1
, it is worth noticing the high number of
missed cycles. In order to minimize it, smaller parts
could be analysed allowing a more sensitive percep-
tion of the f
0
temporal evolution. However, sensi-
tivity to noise presence is also augmented, producing
poorly results when determining the cycles size.
For the ACC signals only the meanwave distance
resulted in high algorithm performance. These results
are possibly related to the higher sensitivity of the
meanwave distance measures associated with the con-
struction of a different meanwave for each part when
the signal is divided into parts.
Analysing the results globally, the L
1
and L
2
dis-
tances returned a total of 571 and 621 errors out of
24982 cycles, achieving 97.7% and 97.5% of accu-
racy, respectively. Besides, the meanwave distance
returned a total of 626 errors out of 25951 cycles,
achieving 97.6% of accuracy.
4 CONCLUSIONS AND FUTURE
WORK
In this paper we presented a signal-independent algo-
rithm for long-term signals processing and time series
clustering. First, an events detection step is taken and
then clustering techniques are applied using a parallel
version of the k-means clustering algorithm capable
of classifying large sized data, obtaining an annotated
signal as output.
In the future, we aim to automatically find the op-
timal length of each part of the divided signal that al-
lows a better monitoring of the temporal evolution of
the fundamental frequency. This would lead to a sig-
nificant reduction in the number of missed cycles.
ACKNOWLEDGEMENTS
This work was partially supported by National
Strategic Reference Framework (NSRF-QREN) un-
der projects AAL4ALL and wiCardioResp, whose
support the authors gratefully acknowledge.
REFERENCES
Chan, F., Yang, Y., Lam, F., Zhang, Y., and Parker, P.
(2000). Fuzzy EMG classification for prosthesis con-
trol. Rehabilitation Engineering, IEEE Transactions
on, 8(3):305–311.
Cuesta-Frau, D., P´erez-Cort´es, J., Andreu-Garc´ıa, G., and
Nov´ak, D. (2002). Feature extraction methods ap-
plied to the clustering of electrocardiographic signals.
A comparative study. In Pattern Recognition, 2002.
Proceedings. 16th International Conference on, vol-
ume 3, pages 961–964. IEEE.
Hansen, P. and Jaumard, B. (1997). Cluster analysis and
mathematical programming. Mathematical program-
ming, 79(1):191–215.
Nunes, N., Ara´ujo, T., and Gamboa, H. (2011). Two-modes
cyclic biosignal clustering based on time series analy-
sis.
PLUX (2012). PLUX - Wireless Biosignals, S.A.
http://www.plux.info/. [Accessed on August, 2012].
Warren Liao, T. (2005). Clustering of time series dataa sur-
vey. Pattern Recognition, 38(11):1857–1874.
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
326
