
Detection of Sharp Wave Activity in Biological Signals using
Differentiation between Consecutive Samples
José L. Ferreira
1
, Pierre J. M. Cluitmans
1,2
and Ronald M. Aarts
1,3
1
Department of Electrical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands
2
Kempenhaeghe Epilepsy Center, Heeze, The Netherlands
3
Philips Research Laboratories Eindhoven, Eindhoven, The Netherlands
Keywords: Sharp Wave Activity Detection, Signal Slope Adaption, Electrocardiogram, QRS Detection.
Abstract: A number of signal processing techniques make use of first-derivative-based approaches for detecting
regions of interest in biological signals. For instance, central and five-point derivative-based algorithms are
employed for emphasizing and identification of the QRS complex in the ECG signal. Signal differentiation
approaches are also used for detection and removal of high-frequency components associated to artefacts in
the EEG signal. This paper aims to present a first-derivative approach based upon differentiation of
consecutive samples – signal slope adaption (SSD) – for detecting regions of sharp wave activity in
biological signals. A case study is analysed whereby SSD is used to mark and select the sharp wave activity
associated to the QRS complex in the electrocardiogram. Evaluation of our methodology reveals that SSD
shows to be effective for identification of QRS samples and, thereby, could be also employed to detect
samples associated to sharp wave activity regions of other biological signals which possess similar signal
slope behaviour.
1 INTRODUCTION
In research and clinical practice, automatic
measurement and recognition of parameters in
biological signals are fundamental to implement
computer-based tools for data analysis and patient
monitoring. In this context, detection of sharp wave
activity or instantaneous signal variability represents
a useful parameter in digital biological signal
processing. A classical example of identification of a
region with steep wave activity in biological signals
is the detection of the QRS complex for
measurement of the heart rate variability (HRV),
which constitutes an important method for
assessment of the cardiac regulation and diagnostic
of disorders such as arrhythmias and congestive
heart failure (Clifford, 2006); (Rangayyan, 2002).
A number of methodologies which make use of
first-derivative-based approaches and differentiation
of the ECG signal are proposed in the literature for
QRS detection (Pan and Tompkins, 1985);
(Hamilton and Tompkins, 1986); (Benitez et al.,
2000); (Köhler et al., 2002); (Rezk et al. 2011). The
basic idea of differentiating the digital signal is that
such a feature can be used for characterizing and
emphasizing regions of the signal which contain
sharp wave activity or specific slope features, as is
the case of the QRS complex (Köhler et al., 2002).
As mentioned by Rangayyan (2002), the QRS
complex has the largest signal slope in a cardiac
cycle due to rapid conduction and depolarization
characteristics of the ventricles. First-derivative
approaches are reported to be robust under
conditions of changes in QRS amplitude and for
ECG excerpts corrupted by baseline drifts, motion
artefacts, and muscular activity (Arzeno et al. 2008);
(Rangayyan, 2002); (Clifford, 2006).
Another first-derivative-based approach for fast
wave activity detection is associated with the
identification and removal of artefacts from the EEG
signal, as proposed by Van de Velde et al. (1998)
and Ferreira et al. (2012). According to Van de
Velde et al. (1998), a slope differentiator procedure
is employed for detection of the larger signal slope
related to the higher-frequency of muscles artefacts
components in comparison to the EEG. By using the
same idea, Ferreira et al. (2012) present an approach
based upon differentiation of consecutive samples of
the EEG signal for identification and removal of
gradient artefacts residuals from the
327
Ferreira J., J. M. Cluitmans P. and M. Aarts R..
Detection of Sharp Wave Activity in Biological Signals using Differentiation between Consecutive Samples.
DOI: 10.5220/0004245003270332
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2013), pages 327-332
ISBN: 978-989-8565-36-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

electroencephalogram recorded within the fMRI
magnetic scanner. Thus, in both of the cases above,
the larger slope of the artefact interference is used to
identify whether or not a sample is artefact free.
First-derivative-based methods have the
advantage of not requiring manual segmentation of
data, training of the algorithms or patient-specific
modifications. Furthermore, they are often
implemented in real-time applications since they do
not require extensive computations (Arzeno et al.,
2008). Signal first-derivative-based approaches can
be used for identifying determined frequency
properties in the signal as well (Cluitmans et al.,
1993); (Van de Velde et al., 1998).
2 OBJECTIVES
Allen et al. (1998) propose a methodology for
ballistogram artefacts removal from the EEG signal
recorded during combined EEG-fMRI which makes
use of R-peaks detection in the ECG signal
simultaneously registered. The ballistogram or pulse
artefact is induced in the electrodes of the
electroencephalograph by the pulsatile movement of
blood in scalp arteries within the magnetic static
field (B
0
) of the magnetic scanner. According to
Allen et al., the identified QRS peaks in the ECG
signal are used to calculate an average pulse artefact
in the EEG signal which is then subtracted from
those regions where the ballistogram artefact
appears. A procedure to extract the ECG peaks
based upon data segmentation and training is
proposed within the methodology for average pulse
artefact subtraction by those authors.
During application of our method proposed in
Ferreira et al. (2012) for identification and removal
of residual gradient artefacts from the EEG signal,
we noticed that the same approach could be
modified and used for detection of the sharp wave
activity associated to the ECG peaks, as well as to
other types of biological signals. Thereby, it could
be used during removal of the ballistogram artefact
according to the methodology of Allen et al. (1998).
Moreover, the advantages mentioned by Arzeno et
al. (2008) by using first-derivative techniques could
be incorporated to that methodology.
The objective of this paper is to propose and
assess an approach for sharp wave identification in
biological signals which makes use of the difference
between consecutive samples of the signal, modified
from Ferreira et al. (2012). In this sense, a case study
is presented in which our method is applied to
identify the sharp wave activity associated to the
QRS complex of the ECG signal.
3 MATERIALS AND METHODS
3.1 Subjects
For application and evaluation of the proposed
methodology for fast wave activity detection, we
used data from the MIT-BH Arrhythmia and the
MIT-BH Noise Stress Test databases (MIT-BIH,
1998). These databases consist of 30 min
ambulatory ECG recordings whose sampling rating
for signal acquiring was 360 samples per second.
For performance evaluation purposes, we
implemented and applied a QRS detector using our
methodology to the 12 recordings of the MIT-BH
Noise Stress Test Database.
3.2 Differentiation between
Consecutive Samples for Sharp
Wave Activity Detection
Ferreira et al. (2012) describe a methodology for
identification and removal of gradient artefact
residuals from the EEG signal which is based upon
the differentiation of consecutive samples of the
digital signal. According to such an approach, the
larger slope associated to the sharp wave activity of
the gradient artefact residuals is used for detecting
EEG samples which contain artefact interference. In
order to identify which samples are in the region of
fast wave activity, a slope threshold (thrs) is
estimated in such a way that if the sample has signal
slope larger than this threshold, it is then classified
to belong to the sharp wave activity region. thrs can
be estimated, for example, taking into account the
probability distribution of the signal slope.
The same idea can be applied for other types of
biological signal with regions of sharp wave activity.
It is the case of the QRS complex whose samples
have signal slope much larger than other regions of
the ECG signal (Rangayyan, 2002). The signal of
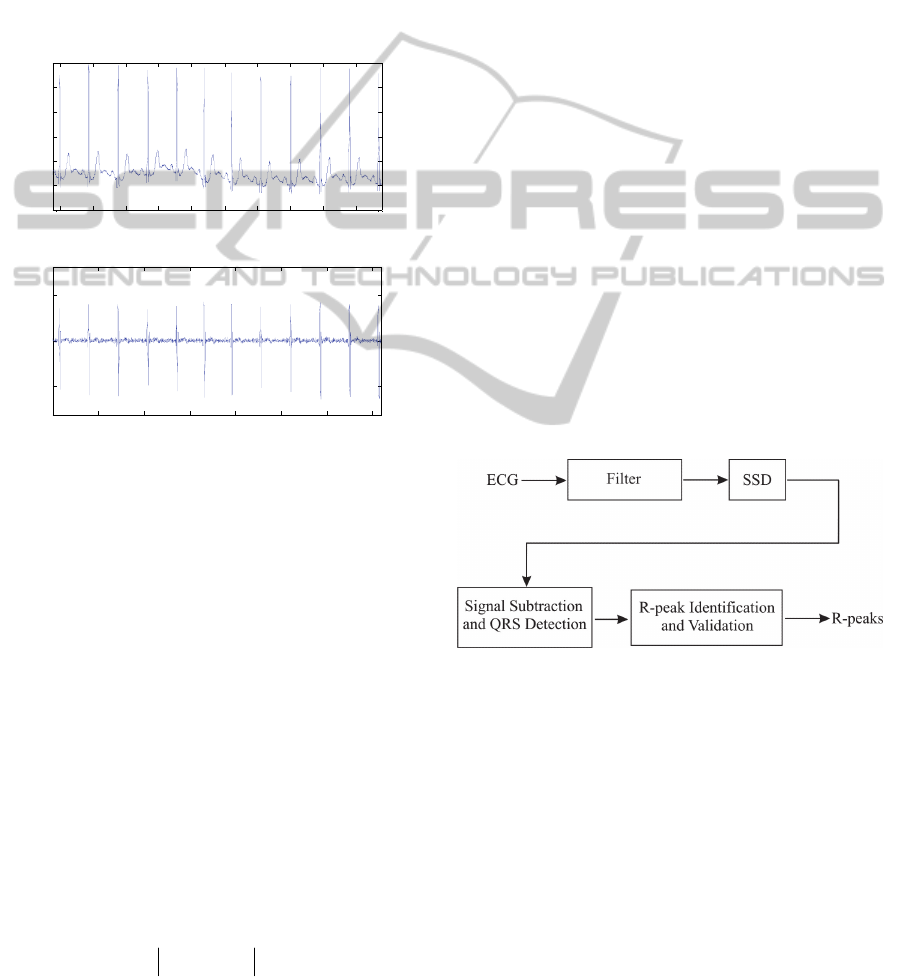
figure 1a consists of an excerpt of 3600 samples
(10 s) of the MIT-BH Arrhythmia Database
recording 103. The respective differentiated signal is
shown in figure 1b. Such a differentiation was
obtained by subtraction of consecutive samples of
the ECG signal, diff (ECG). Clearly, it can be
noticed that higher values of the differentiated ECG
are coincident with the region of the QRS complex.
By analysing probability distributions of the
signal slope of standard clinical ECG signals with
high SNR, we could infer that the slope of samples
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
328
which belong to QRS regions are located above the
threshold, thrs, calculated considering the average
and the standard deviation of diff (ECG):
)(
)(diff)(diff
σμthrs
ECGECG
,
(1)
where μ
diff (ECG)
is the average and σ
diff (ECG)
is the
standard deviation of diff (ECG), considering a
window whose number of points is equal to the
length of the differentiated ECG. It is noteworthy
that the parameter thrs also corresponds to the RMS
value of diff (ECG).
808 809 810 811 812 813 814 815 816 817
-1
-0.5
0
0.5
1
1.5
2
ECG signal
Signal (
V)
Tim e(s)
0 500 1000 1500 2000 2500 3000 350
0
-0.5
0
0.5
Sam
p
le
Diff (ECG) (
V/sample)
Differentiated ECG signal
a
b
Figure 1: (a) Excerpt of 3600 samples (10 s) of the MIT-
BH Arrhythmia Database recording 103 and (b) respective
differentiated signal diff (ECG).
Taking into account the remarks above, a
modified methodology from Ferreira et al. (2012),
herein named signal slope adaption (SSD), was
developed and is employed in this work in order to
carry out the localization of the sharp wave activity
associated to the QRS complexes, described as
follows.
As mentioned earlier, the highest values of the
signal differentiation, diff (ECG), occurs precisely
for samples located in the QRS complex. Therefore,
once the highest slopes of the ECG signal can be
associated to QRS samples along the ECG signal,
they could be adequately identified.
The maximum absolute value of the difference
between consecutive samples of the ECG signal
corresponds to the parameter r
i
which is related to
diff (ECG) by the following expression:
max
i
r diff ECG
,
(2)
where i is the subscript of the maximum slope within
diff (ECG).The two consecutive samples ECG
i
and
ECG
i+1
associated to r
i
are adapted by using (3):
iii,corct
LECGECG
,
iii,corct
LECGECG
11
,
(3)
where
thrsrL
ii
.
(4)
In (3), the sign of L
i
is set positive when
ECG
i
> ECG
i+1
, and vice-versa.
The signal ECG in equation (2) is then replaced
by the modified signal ECG
corct
which contains the
adapted samples ECG
corct,i
and ECG
corct,i+1
. (2), (3),
and (4) are iteratively recalculated until L
i
≤ 0. The
decreasing value of r
i
calculated at each iteration
ensures the convergence of the parameter L
i
. After
the last iteration, all samples of ECG which have
slope larger than thrs are adapted within ECG
corct
and, therefore, match the samples of the QRS
complex.
3.3 QRS Detector for Methodology
Evaluation
The following QRS detector was implemented for
evaluation of our methodology, according to the
common algorithm structure proposed for QRS
detection (Köhler et al., 2002):
Figure 2: Block diagram structure of the QRS detector
algorithm for methodology evaluation.
As observed in the diagram above, a filter is
applied to the ECG signal before application of SSD.
As is done with most QRS detector algorithms, a
band-pass filter was used. This filter was set up as a
56-coefficient FIR, cut-off frequencies at 8 and
35 Hz. The reason to set the cut-off frequency at
35 Hz is because we noticed that a lower value
causes considerable attenuation of the amplitude of
QRS samples. This fact is in agreement with Thakor
et al. (1984) which indicate that the bandwidth of the
QRS complex ranges from 5 to around 40 Hz.
Application of the filter stage of figure 2 could
be bypassed when the ECG signal is affected by
DetectionofSharpWaveActivityinBiologicalSignalsusingDifferentiationbetweenConsecutiveSamples
329

artefacts whose signal slope has order of magnitude
much lower than the slope of the QRS samples, as
discussed below.
3.4 Signal Subtraction and QRS
Detection
As depicted in figure 2, after SSD approach
described by (1) – (4) to be employed for adapting
the samples associated to the QRS complex, a
further subtraction stage is carried out for QRS
samples selection, as follows:
corctfilt
ECGECGP
sig
,
(5)
where P
sig
contains the QRS selected samples.
ECG
filt
and ECG
corct
correspond to the ECG signal
after filtering and SSD application respectively. As
SSD approach adapts samples with larger signal
slopes associated to the QRS complex, the
subtraction indicated in (5) excludes other regions of
the ECG signal in such a way that the latter are
represented as zero values within P
sig
. Therefore, the
samples of P
sig
whose value is different from zero
are assumed to belong to the QRS complex.
3.5 R-peak Identification and
Validation
Motion artefacts and drifting baselines are corrected
during the subtraction performed in (5). Hence, the
filtering stage shown in figure 2 could be used only
when the signal is affected by artefacts whose signal
slope is higher than thrs. Otherwise, there is no need
for calculating another threshold for identification of
the R-peaks as well. In this situation, since the
samples of P
sig
could be grouped in clusters
corresponding to each QRS complex, the maximum
sample amplitude of each cluster corresponds to the
respective R-peak.
On the other hand, R-peak validation rules are
demanded when the noise sample slope is higher
than thrs, and calculation of a second threshold is
also necessary. We tested a second threshold, trp,
calculated taking into account the RMS value of the
samples that belong to clusters corresponding to the
last QRS complexes located. trp was set as being
50% of such a RMS value.
Also for peak validation, the minimum time
between two consecutive clusters was set as 200 ms,
considering the ECG refractory period. Thus, when
two consecutive clusters along P
sig
had a time
difference lower than 200 ms, they were grouped
into a unique cluster whose maximum sample
amplitude was validated as identified R-peak.
3.6 QRS Detection Performance
Analysis
According to Köhler et al. (2002), the usage of
software QRS detection algorithms requires the
evaluation of the detection performance. In this way,
ANSI/AAMI/ISO EC57 (1998) recommends that the
parameters sensitivity (Se) and positive predictivity
(+P) should be calculated for algorithm assessment:
FNTP
TP
Se
,
(6)
FPTP
TP
P
,
(7)
where TP is the number of true positives, FN the
number of false negatives, and FP is the number of
false positive QRS predictions.
4 RESULTS
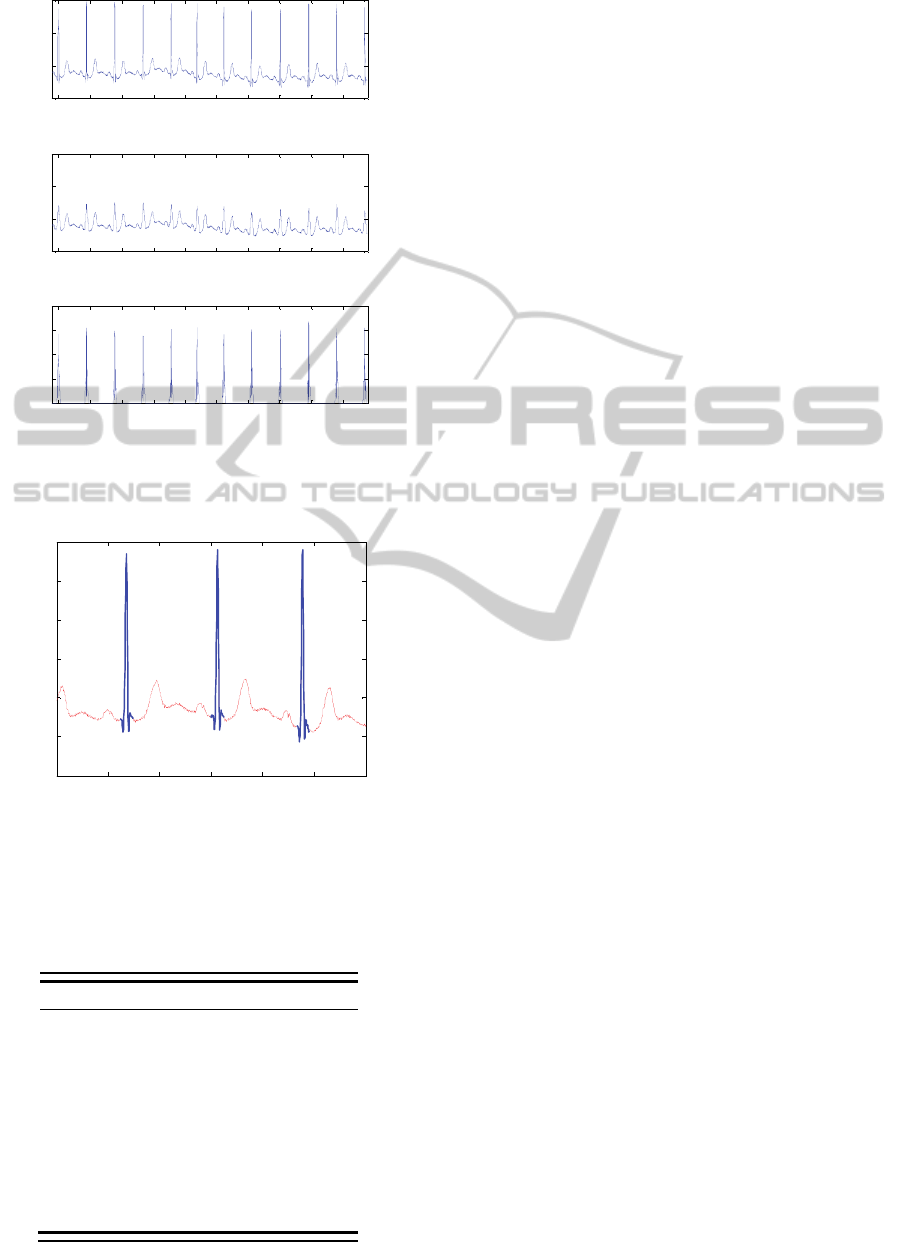
Figure 3 illustrates the application of our
methodology in the ECG excerpt of figure 1a.
For the signal shown in figure 3a, the band-pass
filter was not applied in order to illustrate the
application of our methodology to a raw ECG signal
with high SNR. As observed in figure 3b, SSD
approach adapts only ECG samples associated to the
sharp wave activity of the QRS complexes whose
slope is higher than thrs. Figure 4 depicts a zooming
in around the time 811.5 s showing the samples
identified as QRS samples.
Evaluation of our methodology was performed
by application of the QRS detector of figure 2 to the
recordings of the MIT-BH Noise Stress Test
Database in accordance with ANSI/AAMI/ISO
EC57 (1998). This database corresponds to twelve
30 min ECG recordings with different levels of SNR
at 0, 6, 12, 18, 24, and -6 dB.
The results obtained for the parameters Se and
+P are presented in table 1. The QRS detector
shows high sensitivity (above 84%) even when the
SNR is about -6 dB. Although the values obtained
for +P are affected by a larger number of false
positives which occur under low SNR conditions,
the noise tolerance performance of such a detector is
comparable to other ones proposed in the literature
(Benitez et al., 2000); (Rezk et al., 2011).
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
330
808 809 810 811 812 813 814 815 816 817
-1
0
1
2
ECG signal
Signal (
V)
Time(s)
808 809 810 811 812 813 814 815 816 817
-1
0
1
2
ECG signal after application of SSD
Signal (
V)
Time(s)
808 809 810 811 812 813 814 815 816 817
0
0.5
1
1.5
2
ECG signal after subtraction stage
Signal
(
V)
Time(s)
a
b
c
Figure 3: (a) ECG signal of figure 1a; (b) ECG
corct
,
resulting from application of SSD to (a); (c) P
sig
or
absolute value of the subtraction between (a) and (b).
810 810.5 811 811.5 812 812.5 813
-1
-0.5
0
0.5
1
1.5
2
Identified QRS samples
Signal (
V)
Time(s)
Figure 4: Zooming in figure 3a, around time 811.5 s. The
regions of sharp wave activity with signal slope higher
than thrs are identified as QRS samples (thick blue traces).
Table 1: Se and +P calculated for the MIT-BH Noise
Stress Test Database considering application of the QRS
detector depicted in figure 2.
Recording
Se (%) +P (%)
118e00 91.62 79.33
118e06 98.24 86.24
118e12 99.96 93.47
118e18 99.96 98.91
118e24 99.96 99.56
118e_6 84.11 74.64
119e00 95.36 82.04
119e06 99.09 87.77
119e12 99.90 97.45
119e18 100 100
119e24 100 100
119e_6 85.51 71.96
5 DISCUSSION
As observed in figure 1, the difference between
consecutive samples of the ECG signal can be used
for identification of the shaper wave activity
associated to QRS complex (Rangayyan, 2002). This
property can be also observed in other biological
signals or under specific conditions where the signal
is affected by artefacts or other types of interference,
as discussed by Cluitmans et al. (1993), Van de
Velde et al. (1998), and Ferreira et al. (2012).
Ferreira et al. propose a methodology for
detection and removal of gradient artefact residuals
which also identifies artefact samples by the
magnitude of the signal slope. Thus, we developed a
modification of such a method for identification and
selection of signal samples which contain sharp
wave activity, described by equations (1) – (4).
When applied to the ECG signal, these equations
results in an effective identification of the QRS
samples, as depicted in figures 3 and 4.
Application of our methodology within the
prototype of QRS detector (figure 2) reveals that it
has a good performance for identifying the R-peaks,
even under conditions of low SNR (table 1).
According to Arzeno et al. (2008), first-derivative-
based methods can be easily implemented in real-
time R-peak detection. Such advantage is also
observed for the detector of figure 2. Moreover, it
shows to be effective for detection of ectopic beats
as well. Hence, in future work the performance of
this QRS detector will be evaluated for a larger set
of data and during removal of the ballistogram
artefact as well, according to the approach proposed
by Allen et al. (1998).
Therefore, the results shown in figures 3 and 4,
and table 1 reflect the effectiveness of SSD in the
detection of the steep wave activity of the QRS
complex. This fact clearly indicates the possibility to
apply the same approach to detect regions or
artefacts in other biological signals which possess
similar behaviour of the slope parameter.
Identification of samples as belonging to the sharp
wave activity region of interest depends on the value
of the slope signal threshold, estimated by equation
(1) for the ECG signal. Thus, our methodology
achieves better performance when the slope of the
sharp wave activity samples is higher and does not
overlap the slope magnitude of other regions of the
analysed signal.
Another fact which should be investigated in
future work is how the application of SSD approach
could be used to identify samples from a specific
frequency bandwidth in the biological signal. For
DetectionofSharpWaveActivityinBiologicalSignalsusingDifferentiationbetweenConsecutiveSamples
331

example, in the case of the QRS complex region,
this bandwidth ranges from around 5 to 40 Hz. In
experiments which involve removal of gradient
artefacts residuals from EEG signals, SSD shows to
select higher frequency components associated to
the artefact (Ritter et al., 2010; Ferreira et al. 2012).
Thereby, SSD could be evaluated and proposed as
an alternative time-domain filtering approach.
6 CONCLUSIONS
In this work, we propose a methodology (SSD) for
detection of sharp wave activity in biological signals
based upon differentiation of consecutive samples of
the digital signal.
Our methodology shows to achieve effective
identification of the sharp wave activity associated
to the QRS samples in the ECG signal. Also
evaluation of a QRS detector prototype which makes
use of SSD reveals that the QRS complexes are
localized with sensitivity and positive predictivity
comparable to other methodologies proposed in the
literature. In future work, our methodology shall be
applied and evaluated for detection of sharp wave
activity in other types of biological signals.
ACKNOWLEDGEMENTS
This work has been made possible by a grant from
the European Union and Erasmus Mundus – EBW II
Project, and by a grant from CNPq – Science
without Borders Program.
REFERENCES
Allen, P., Polizzi, G., Krakow, K., Fish, D., Lemieux, L.,
1998. Identification of EEG events in the MR scanner:
the problem of pulse artefact and a method for its
subtraction. NeuroImage. 8, 229-239.
ANSI/AAMI/ISO EC57, 1998. Testing and reporting
performance results of cardiac rhythm and ST segment
measurement algorithms. AAMI Recommended
Practice/ American National Standard.
Arzeno, N., Deng, Z., Poon, C., 2008. Analysis of first-
derivative based QRS detection algorithms. IEEE
Trans. Biomed. Eng. 55 (2), 478-484.
Benitez, D., Gaydecki, P., Zaidi, A., Fitzpatrick, A., 2000.
A new QRS detection algorithm based on the Hilbert
transform. Comput. Cardiol. 27, 379-382.
Clifford, G., 2006. ECG statistics, noise, artifacts, and
missing data. In G. Clifford, F. Azuaje, P. McSharry,
(eds.), Advanced tools for ECG data analysis. Artech
House: Boston, London.
Cluitmans, P., Jansen, J., Beneken, J., 1993. Artefact
detection and removal during auditory evoked
potential monitoring. J. Clin. Monit. 9 (2), 112–120.
Ferreira, J., Cluitmans, P., Aarts, R. M., 2012. Gradient
artefact correction in the EEG signal recorded within
the fMRI scanner. Proceedings of the 5
th
International
Conference on Bio-inspired Systems and Signal
Processing, BIOSIGNALS 2012, Vilamoura, Portugal,
February 1 – 4, 2012. 110-117.
Hamilton, P., Tompkins, W., 1986. Quantitative
investigation of QRS detection rules using the
MIT/BIH Arrhythmia Database. IEEE Trans. Biomed.
Eng. 33 (12), 1157-1165.
Köhler, B., Hennig, C., Orglmeister, R., 2002. The
principles of software QRS detection. IEEE Eng. Med.
Biol. Mag. 21 (1), 42-57.
MIT-BIH, 1998. Database Distribution. Massachusetts
Institute of Technology, Cambridge, MA. Available:
http://ecg.mit.edu/.
Pan, J., Tompkins, W., 1985. A real-time QRS detection
algorithm. IEEE Trans. Biomed. Eng. 32 (3), 230-236.
Rangayyan, R., 2002. Biomedical signal analysis: a case-
study approach. Wiley: New York.
Rezk, S., Join, C., Asmi, S., 2011. An algebraic
derivative-based method for R wave detection.
Proceedings of the 19
th
European Signal Processing
Conference, EUSIPCO 2011, Barcelona, Spain,
August 29 – September 2, 2011. 1578-1582.
Ritter, P., Becker, R., Freyer, F., Villringer, A., 2010. EEG
quality: the image acquisition artifact. In C. Mulert, L.
Limieux (eds.), EEG-fMRI: Physiological basis,
technique and applications. Springer: Verlag, Berlin,
Heidelberg.
Thakor, N., Webster, J., Tompkins, W., 1984. Estimation
of QRS complex power spectra for design a QRS
filter. IEEE Trans. Biomed. Eng. 31 (11), 702-706.
Van de Velde, R., Van Erp, G., Cluitmans, P., 1998.
Detection of muscle artefact in the normal human
awake EEG. Electroencephalogr. Clin. Neurophysiol.
107, 149-158.
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
332
