
Na
¨
ıve Bayes Domain Adaptation for Biological Sequences
Nic Herndon and Doina Caragea
Computing and Information Sciences, Kansas State University, 234 Nichols Hall, 66506, Manhattan, KS, U.S.A.
Keywords:
Na
¨
ıve Bayes, Domain Adaptation, Supervised Learning, Semi-supervised Learning, Self-training, Biological
Sequences, Protein Localization.
Abstract:
The increased volume of biological data requires automatic computation tools to analyze it. Although ma-
chine learning methods have been successfully used with biological sequences in a supervised framework,
their accuracy usually suffers when a classifier is learned on a source domain and applied to a different, less
studied domain, in a domain adaptation framework. To address this issue, we propose to use an algorithm
that combines labeled sequences from a well studied organism, the source domain, with labeled and unlabeled
sequences from a related, less studied organism, the target domain. Our experimental results show that this
algorithm has high classifying accuracy on the target domain.
1 INTRODUCTION
The widespread use of next generation sequencing
(NGS) technologies in the recent years has resulted
in an increase in the volume of biological data gener-
ated, including both DNA sequences and also derived
protein sequences. A challenge arising from the in-
creased volume of data consists of the organization,
analysis, and interpretation of this data, in order to
create or improve genome assemblies or genome an-
notation, or to predict protein function, structure and
localization, among others. Some of these problems
can be framed as biological sequence classification
problems, i.e., assigning one of several labels to a
DNA or protein sequence based on its content (e.g.,
predicting the presence or absence of an acceptor or
donor splice site in DNA sequences centered around
GT or AG dimers; or determining where a protein
is localized, such as in cytoplasm, inner membrane,
periplasm, outer membrane, or extracellular space,
a.k.a., protein localization).
Using machine learning or statistical inference
methods allows labeling of biological data several or-
ders of magnitude faster than it can be done manu-
ally, and with high accuracy. For example, hidden
Markov models are currently used in gene prediction
algorithms, and support vector machines have shown
promising results with handwritten digit classification
(Vapnik, 1995), optical character recognition (M
¨
uller
et al., 2001; Sch
¨
olkopf and Smola, 2001) and transla-
tion initiation sites classification based on proximity
to start codon within sequence window (M
¨
uller et al.,
2001) or based on positional nucleotide incidences
(Zien et al., 2000), classification into malign or be-
nign of gene expression profiles (Noble, 2006), ab ini-
tio gene prediction (Bernal et al., 2007), classification
of DNA sequences into sequences with splice site at
a determined location or not (Jaakkola and Haussler,
1999; Sonnenburg et al., 2007; Tsuda et al., 2002;
Sonnenburg et al., 2002; Lorena and de Carvalho,
2003; R
¨
atsch and Sonnenburg, 2004; Degroeve et al.,
2005; Huang et al., 2006; Zhang et al., 2006; Baten
et al., 2006), and classifying the function of genes
based on gene expression data (Brown et al., 2000).
However, using a supervised classifier trained on
a source domain to predict data on a different target
domain usually results in reduced classification ac-
curacy. Instead of using the supervised classifier, an
algorithm developed in the domain adaptation frame-
work can be employed to transfer knowledge from the
source domain to the target domain. Such an algo-
rithm has to take into consideration the fact that some,
if not all, of the features have different probabilities in
the target and source domains (Jiang and Zhai, 2007).
In other words, some of the features that are corre-
lated to a label in the source domain might not be cor-
related to the same or any label in the target domain,
while, some of the features have the same label corre-
lations between the source and target domains. The
former ones are known as domain specific features
and the latter ones are generalizable features (Jiang
and Zhai, 2007).
62
Herndon N. and Caragea D..
Naïve Bayes Domain Adaptation for Biological Sequences.
DOI: 10.5220/0004245500620070
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2013), pages 62-70
ISBN: 978-989-8565-35-8
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

Domain adaptation algorithms are particularly ap-
plicable to many biological problems for which there
is a large corpus of labeled data for some well stud-
ied organisms and much less labeled data for an or-
ganism of interest. Thus, when studying a new or-
ganism, it would be preferable if the knowledge from
other, more extensively studied organism(s), could be
applied to a lesser studied organism. This would al-
leviate the need to manually generate enough labeled
data to use a machine learning algorithm to make pre-
dictions on the biological sequences from the target
domain. Instead, we could filter out the domain spe-
cific features from the source domain and use only the
generalizable features between the source and target
domains, together with the target specific features, to
classify the data.
Towards this goal, we modified the Adapted Na
¨
ıve
Bayes (ANB) algorithm to make it suitable for the
biological data. We chose this algorithm because
Na
¨
ıve Bayes based algorithms are faster and require
no tuning. In addition, this algorithm was success-
fully used by Tan et al. (2009) on text classification
for sentiment analysis, discussed in Section 2. It com-
bines a weighted version of the multinomial Na
¨
ıve
Bayes classifier with the Expectation-Maximization
algorithm. In the maximization step, the class proba-
bilities and the conditional feature probabilities given
the class are calculated using a weighted combina-
tion between the labeled data from the source domain
and the unlabeled data from the target domain. In
the expectation step, the conditional class probabili-
ties given the instance are calculated with the proba-
bility values from the maximization step using Bayes
theorem. The two steps are repeated until the prob-
abilities in the expectation step converge. With each
iteration, the weight is shifted from the source data
to the target data. The key modifications we made
to this algorithm are the use of labeled data from the
target domain, and the incorporation of self-training
(Yarowsky, 1995; Riloff et al., 2003; Maeireizo et al.,
2004) to make it feasible for biological data, as pre-
sented in more detail in Section 3.
We tested the ANB classifier on two biological
datasets, as described in the Section 3.4, for classify-
ing localization of proteins. The experimental results,
Section 3.6, show that this classifier achieves classifi-
cation accuracy than a Na
¨
ıve Bayes classifier trained
on the source domain and tested on the target domain,
especially when the two domains are less related.
2 RELATED WORK
Up to now, most of the work in domain adaptation
tion has been on non-biological problems. For in-
stance, text classification has received a lot of atten-
tion in the domain classification framework. One ex-
ample, the Na
¨
ıve Bayes Transfer Classification algo-
rithm (Dai et al., 2007), assumes that the source and
target data have different distributions. It trains a clas-
sifier on source data and then applies the Expectation-
Maximization (EM) algorithm to fit the classifier for
the target data, using the Kullback-Liebler divergence
to determine the trade-off parameters in the EM al-
gorithm. When tested on datasets from Newsgroups,
SRAA and Reuters for the task of top-category clas-
sification of text documents this algorithm performed
better than support vector machine and Na
¨
ıve Bayes
classifiers.
Another algorithm derived from the Na
¨
ıve Bayes
classifier that uses domain adaptation is the Adapted
Na
¨
ıve Bayes classifier (Tan et al., 2009), which iden-
tifies and uses only the generalizable features from
the source domain, and the unlabeled data with all the
features from the target domain to build a classifier
for the target domain. This algorithm was evaluated
on transferring the sentiment analysis classifier from
a source domain to several target domains. The pre-
diction rate was promising, with Micro F1 values be-
tween 0.69 and 0.90, and Macro F1 values between
0.59 and 0.91. However, the classifier did not use any
labeled data from the target domain.
Nigam et al. (1999) showed empirically that com-
bining a small labeled dataset with a large unlabeled
dataset from the same or different domains can re-
duce the classification error of text documents by up
to 30%. Their algorithm also uses a combination of
Expectation Maximization and the Na
¨
ıve Bayes clas-
sifier by first learning a classifier on the labeled data
which is then used to classify the unlabeled data. The
combination of these datasets trains a new classifier
and iterates until convergence. By augmenting the la-
beled data with unlabeled data the number of labeled
instances was smaller compared to using only labeled
data.
For biological sequences, most domain adaptation
algorithms employed support vector machines. For
example, Sonnenburg et al. (2007) used a Support
Vector Machine with weighted degree kernel to clas-
sify DNA sequences into sequences that have or not
have a splice site at the location of interest. Even
though the training data was highly skewed towards
the negative class, their classifier achieved good ac-
curacy.
For more work on domain adaptation and transfer
learning, see the survey by Pan and Yang (2010) .
NaïveBayesDomainAdaptationforBiologicalSequences
63
3 METHODOLOGY
3.1 Identifying and Selecting
Generalizable Features from the
Source Domain
To successfully adapt a classifier from the source do-
main to the target domain, the classifier has to iden-
tify in the source domain the subset of the features
that generalize well and are highly correlated with the
label. Then, use a combination of only these features
from the source domain and all the features from the
target domain to predict the labels in the target do-
main.
We used the feature selection method proposed by
Tan et al. (2009). Theoretically, the set of features in
each domain can be split into four categories, based
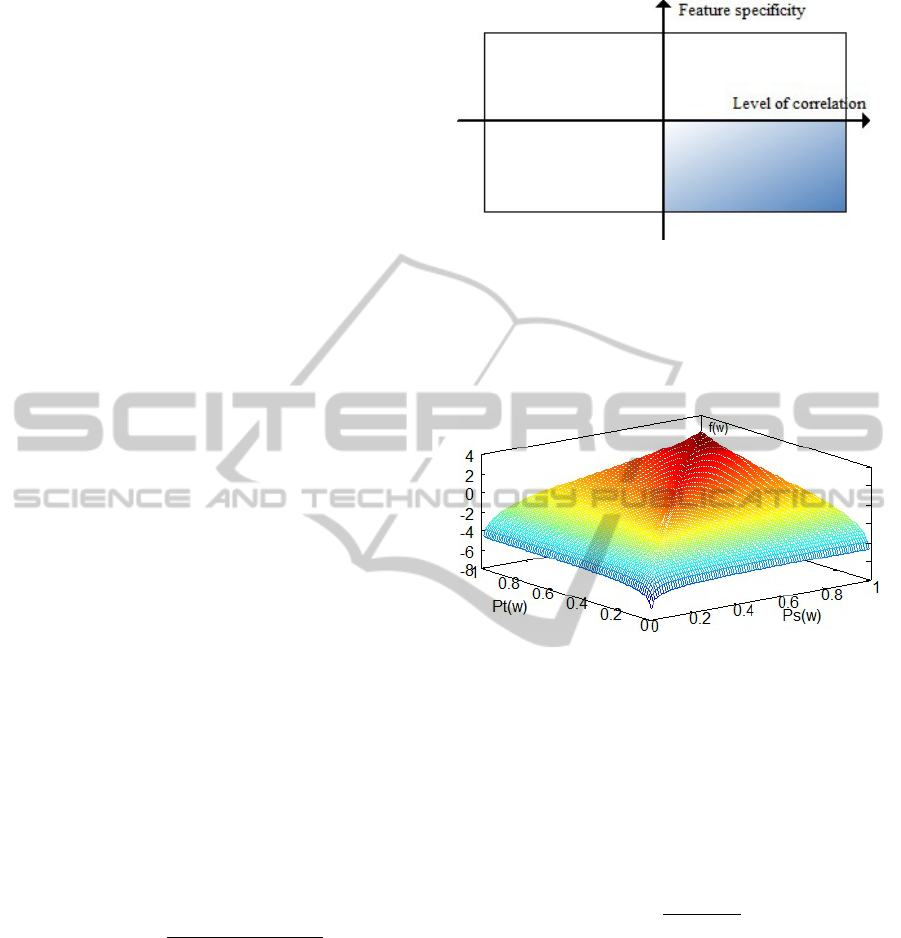
on two selection criteria (Figure 1). The first selec-
tion criterion is the level of correlation. The features
have varying degrees of correlation with the label as-
signed to a sequence. Based on the correlation be-
tween the feature and the label, the features can be
divided into features that are highly related to the la-
bels, and features that are less related to the labels.
The second selection criterion is the specificity of the
features. Based on this criterion, the features can be
divided into features that are very specific to a do-
main, and features that generalize well across related
domains.
To select these features from the source domain
we rank all the features from the source domain based
on their probabilities. The features that are gener-
alizable would most likely occur frequently in both
domains, and should be ranked higher. Moreover,
the features that are correlated to the labels should
be ranked higher, as well (Figure 2). Therefore, we
use the following measure to rank the features in the
source domain:
f (w) = log
P
s
(w) · P
t
(w)
|P
s
(w) − P
t
(w)| + α
(1)
where P
s
and P
t
are the probability of the feature w
in the source and target domain, respectively. The
numerator ranks higher the features that occur fre-
quently in both domains, since the larger both prob-
abilities are the larger the numerator is, and thus the
higher the rank of the feature is. The denominator
ranks higher the features that have similar probabili-
ties (i.e., the generalizable features), since the closer
the probabilities are for a feature in both domains, the
smaller the denominator value is, and thus the higher
the rank. The additional value in the denominator, α,
is used to prevent division by zero. The higher its
Figure 1: Feature selection. Based on the correlation with
the label, the features can be split into features weakly cor-
related with the label (left) and features highly correlated
with the label (right). Based on how specific the features
are, they can be split into domain specific features (top) and
generalizable features (bottom). Our goal is to select the
features in the bottom-right quadrant.
Figure 2: Ranking of features in the source domain using
Equation (1). The rank of a feature is higher if it has a high
probability or occurs with similar probability in the target
domain. Note: This graph was drawn using Octave (Eaton
et al., 2008).
value is the more influence the numerator has in rank-
ing the features, and vice versa. To limit its influence
on ranking the features, we chose a small value for
this parameter, 0.0001. The probability of a feature in
either domain is
P(w) =
N(w) + β
|D| + 2 ·β
(2)
where N is the number of instances in the domain in
which the feature w occurs, D is the total number of
instances in the domain and β is a smoothing factor,
which is used to prevent the probability of a feature
to be 0 (which would make the numerator in (1) equal
to 0, and the logarithm function is undefined for 0).
We chose a small value for β as well, 0.0001, to limit
its influence on the ranking of features. Note that the
values for α and β do not have to be the same, but they
can be, as used by Tan et al. (2009) and in our case.
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
64

3.2 Multinomial Na
¨
ıve Bayes (MNB)
Classifier
The multinomial na
¨
ıve Bayes classifier (Mccallum
and Nigam, 1998) assumes that the sample data used
to train the classifier is representative of the popula-
tion data on which the classifier will be used. In ad-
dition, it assumes that the frequency of the features
determines the label assigned to an instance, and that
the position of a feature is irrelevant (the na
¨
ıve Bayes
assumption). Thus, using Bayes’ property a classi-
fier can approximate the posterior probability, i.e., the
probability of a class given an unclassified instance,
as being proportional to the product of the prior prob-
ability of the class, and the feature conditional proba-
bilities given an instance from the sample data:
P(c
k
| d
i
) ∝ P(c
k
)
∏
t∈|V |
[P(w
t
| c
k
)]
N
t,i
(3)
where the probability of the class is
P(c
k
) =
∑
i∈|D|
P(c
k
| d
i
)
|D|
(4)
and the conditional probability is
P(w
t
| c
k
) =
∑
i∈|D|
N
t,i
· P(c
k
| d
i
) + 1
∑
t∈|V |
∑
i∈|D|
N
t,i
· P(c
k
| d
i
) + |V |
(5)
Here, N
t,i
is the number of times feature w
t
occurs in
instance d
i
, |V | is the number of features, and |D| is
the number of instances.
3.3 Adapted Na
¨
ıve Bayes Classifier for
Biological Sequences
One limitation of the MNB classifier is that it can only
be trained on one domain, and when the trained clas-
sifier is used on a different domain, in most cases, its
classification accuracy decreases. To address this, we
used the Adapted Na
¨
ıve Bayes (ANB) classifier pro-
posed by Tan et al. (2009), with two modifications:
we used the labeled data from the target domain, and
employed the self-training technique. These will be
described in more detail shortly.
The ANB algorithm is a combination of the
expectation-maximization (EM) algorithm and a
weighted multinomial Na
¨
ıve Bayes algorithm. Sim-
ilar to the EM algorithm, it has two steps that are
iterated until convergence. In the first step, the M-
step, we simultaneously calculate the class probabil-
ity and the class conditional probability of a feature.
However, unlike the EM algorithm that uses the data
from one domain to calculate these values, this algo-
rithm uses a weighted combination of the data from
the source domain and the target domain.
P(c
k
) =
(1 − λ)
∑
i∈D
s
P(c
k
| d
i
) + λ
∑
i∈D
t
P(c
k
| d
i
)
(1 − λ)|D
s
| + λ|D
t
|
(6)
P(w
t
| c
k
) =
(1 − λ)(η
t
N
s
t,k
) + λN
t
t,k
+ 1
(1 − λ)
∑
t∈|V|
η
t
N
s
t,k
+ λ
∑
t∈|V|
N
t
t,k
+ 1
(7)
where N
t,k
is the number of times feature w
t
occurs in
a domain in instances labeled with class k:
N
t,k
=
∑
i∈D
N
t,i
P(c
k
| d
i
) (8)
λ is the weight factor between the source and target
domains:
λ = min{δ · τ, 1} (9)
and τ is the iteration number, δ ∈ (0, 1) is a constant
that determines how fast the weight shifts from the
source domain to the target domain, and η
t
is 1 if fea-
ture t in the source domain is a generalizable feature,
0 otherwise.
Unlike the algorithm proposed by Tan et al.
(2009), which considers that all the instances from
the target domain are unlabeled and does not use them
during the first iteration (i.e., λ = 0), it is reasonable
to assume that there is a small number of labeled in-
stances in the target domain, and our algorithm uses
any labeled data from the target domain in the first
and subsequent iterations. In the first iteration we use
only labeled instances from the source and target do-
mains to calculate the probability distributions for the
class conditional probabilities given the instance. In
subsequent iterations we use the class of the instance
for the labeled data from the source and target do-
mains and the probability distribution of the class for
the unlabeled data from the target domain.
In the second step, the E-step, we estimate the
probability of the class for each instance with the val-
ues obtained from the M-step.
P(c
k
| d
i
) ∝ P(c
k
)
∏
t∈|V|
[P(w
t
| c
k
)]
N
t,i
(10)
The second modification we made to the ANB
classifier (Tan et al., 2009), is our use of self-training,
i.e., at each iteration, we select, proportional to the
class distribution, the instances with the top class
probability, and consider these to be labeled in the
subsequent iterations. This improves the prediction
accuracy of our classifier because it does not allow
the unlabeled data to alter the class distribution from
the target labeled data.
NaïveBayesDomainAdaptationforBiologicalSequences
65

1 Load the data from the source domain, D
s
, the tar-
get domain, D
t
, and parameters α, β, δ.
2 Select generalizable features from the source do-
main, i.e., the top ranked features using Equation
(1).
3 For each class simultaneously calculate the class
probability and the class conditional probability
of each feature using Equations (6-7). For the
source domain use all labeled instances, and only
the generalizable features. For the target domain
use only labeled instances, and all features.
4 Select, proportional to the class distribution, the
target instances with the top class probability, and
consider these to be labeled in the subsequent it-
erations.
5 Loop until the labels assigned to unlabeled data
don’t change.
5.a M-step. Same as step 3 but use the class for la-
beled and self-trained instances from the target
domain, and the class distribution for unlabeled
instances.
5.b Same as step 4.
5.c E-step. Calculate the class distribution for un-
labeled training instances from the target do-
main using Equation (10).
6 Use classifier to label new target data.
Figure 3: Outline of the Adapted Na
¨
ıve Bayes algorithm for
biological sequences.
The two steps, E and M, are repeated until the
instance conditional probabilities values in (10) con-
verge (or a given number of iterations is reached). The
algorithm is summarized in Figure 3.
3.4 Data Sets
We used two data sets to evaluate our classifier. The
first data set, PSORTb v2.0
1
(Gardy et al., 2005),
was first introduced in (Gardy et al., 2003), and con-
tains proteins from gram-negative and gram-positive
bacteria and their primary localization information:
cytoplasm, inner membrane, periplasm, outer mem-
brane, and extracellular space. For our experiments,
we identified classes that appear in both datasets, and
used 480 proteins from gram-positive bacteria (194
from cytoplasm, 103 from inner membrane, and 183
from extracellular space) and 777 proteins from gram-
1
Downloaded from http://www.psort.org/dataset/
datasetv2.html
negative bacteria (278 from cytoplasm, 309 from in-
ner membrane, and 190 from extracellular space).
The second data set, TargetP
2
, was first introduced
in (Emanuelsson et al., 2000), and contains plant and
non-plant proteins and their subcellular localization:
mitochondrial, chloroplast, secretory pathway, and
“other.” From this data set we used 799 plant pro-
teins (368 mitochondrial, 269 secretory pathway and
162 “other”) and 2,738 non-plant proteins (371 mito-
chondrial, 715 secretory pathway and 1652 “other”).
Predicting protein localization is an important biolog-
ical problem because the function of the proteins is
related to their localization.
3.5 Data Preparation and Experimental
Setup
We represent each sequence as a count of occurrences
of k-mers. We use a sliding window approach to count
the k-mer frequencies. For example, the protein se-
quence LLRSYRS would be transformed when using
2-mers into 1, 1, 2, 1, 1 which are the counts corre-
sponding to the occurrences of features LL, LR, RS,
SY, YR.
In order to obtain unbiased estimates for classi-
fier performance we used five-fold cross validation.
We use all labeled data from the source domain for
training (tSL) and randomly split the target domain
data into 3 sets: 20% used as labeled data for training
(tTL), 60% used as unlabeled data for training (tTU),
and 20% used as test data (TTL). So, we train our
classifier on tSL + tTL + tTU and test it on TTL.
We wanted to answer several questions - specifi-
cally, how does the performance of the classifier vary
with:
Q1 Features used (i.e., 3-mers, 2-mers, or 1-mers)?
Q2 Number of features used in the target domain (i.e.,
keep all features, remove at most 50% of the least
occurring features)?
Q3 Number of features retained in the source domain
after selecting the generalizable features?
Q4 Variation with the size of the target la-
beled/unlabeled data set (i.e., train on 100% tSL
+ x% tTL + y% tTU, where x ∈ {5, 10, 20} and
y ∈ {20, 40, 60})?
Q5 The distance between the source and target do-
mains?
Q6 The choice of the source and target domains?
2
Downloaded from http://www.cbs.dtu.dk/services/
TargetP/datasets/datasets.php
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
66

As baselines, we compared our classifier (ANB)
with the multinomial Na
¨
ıve Bayes classifier trained
on all source data (MNB s), the multinomial Na
¨
ıve
Bayes classifier trained on 5% target data (MNB 5t),
and the multinomial Na
¨
ıve Bayes classifier trained
on 80% target data (MNB 80t). Each classifier was
tested on 20% of target data. The expectation is that
the prediction accuracy of our classifier will be lower
bounded by MNB 5t, upper bounded by MNB 80t,
and be better than MNB s.
To evaluate our classifier we used the area under
the receiver operating characteristic (auROC), as the
class distributions are relatively balanced.
3.6 Results
This section provides empirical evidence that aug-
menting the labeled data from a source domain with
labeled and unlabeled data from the target domain
with the ANB algorithm improves the classification
accuracy compared to using only the limited labeled
data from the target domain or using only the data
from a source domain with the MNB classifier.
Table 1 shows the average auROC values over the
five-fold cross validation trials for our algorithm and
for the baseline algorithms on the two datasets used.
For our algorithm, we used different amounts of la-
beled and unlabeled data from the target domain. For
example, the top-left value is the auROC for our algo-
rithm trained on 5% labeled data and 20% unlabeled
data. In each subtable the largest auROC value for the
ANB is highlighted.
We noted that the performance of the ANB classi-
fier varies, as follows:
A1 The best results were obtained when using 3-mers
as features. This makes sense since longer k-mers
capture more information associated with the rel-
ative position of each amino-acid. When using
3-mers, our algorithm provides between 9.84%
and 34.14% better classification accuracy when
compared to multinomial Na
¨
ıve Bayes classifier
trained on 5% of the labeled data from the tar-
get domain, and between 0.37% and 28.2% when
compared to the multinomial Na
¨
ıve Bayes clas-
sifier trained on labeled data from the source do-
main, except when the plant proteins are the target
domain.
A2 When trying to establish how many features from
the target domain should be used we determined
that removing any features does not improve the
performance of our algorithm.
A3 When trying to ascertain how many features from
the source domain should be kept after ranking
them with Equation 1, we determined that the best
results were obtained when at least 50% of the
features were kept, i.e., the 50% top-ranked fea-
tures and any other features with the same rank as
the last feature kept.
A4 For most cases, the largest auROC values for
our algorithm were obtained when using the least
amount of target unlabeled data. This would sug-
gest that even though using unlabeled data is ben-
eficial, using too much unlabeled data is detrimen-
tal because the unlabeled instances act as noise
and corrupt the prediction from the target labeled
data. In addition, intuitively, using more labeled
data from the target domain should lead to bet-
ter prediction accuracy. This was indeed the case
with our classifier.
A5 When the source and target domains are close the
classifier learned is better. For example, the au-
ROC is higher for the PSORTb datasets than for
the TargetP datasets. Therefore, the closer the tar-
get domain is to the source domain the better the
classifier learned.
A6 For the PSORTb dataset, the ANB classifier
had better prediction accuracy when the gram-
negative proteins were used as the source domain
than when the gram-positive proteins were used
as the source domain. Similarly, for the TargetP
dataset, we obtained better predictions when us-
ing non-plant proteins as the source domain than
when using plant proteins as the source domain.
This is because in both cases there were more
gram-negative instances and more non-plant in-
stances, respectively, than gram-positive instances
and plant instances, respectively.
It is interesting to note that in some instances the
multinomial Na
¨
ıve Bayes classifier trained on the
source domain performed better than our algorithm.
This occurred mainly when our algorithm used 5% or
10% of the target labeled data and when the features
were 1-mers or 2-mers. However, this is somewhat
expected, as using very little labeled data from the tar-
get domain does not provide a representative sample
for the population, and using short k-mers does not
capture the relative position of the amino-acids.
3.7 Preliminary Results on a Third
Dataset
We have also done a preliminary analysis on a third
data set
3
, first introduced in (Schweikert et al., 2008).
3
Downloaded from ftp://ftp.tuebingen.mpg.de/fml/
cwidmer/
NaïveBayesDomainAdaptationforBiologicalSequences
67
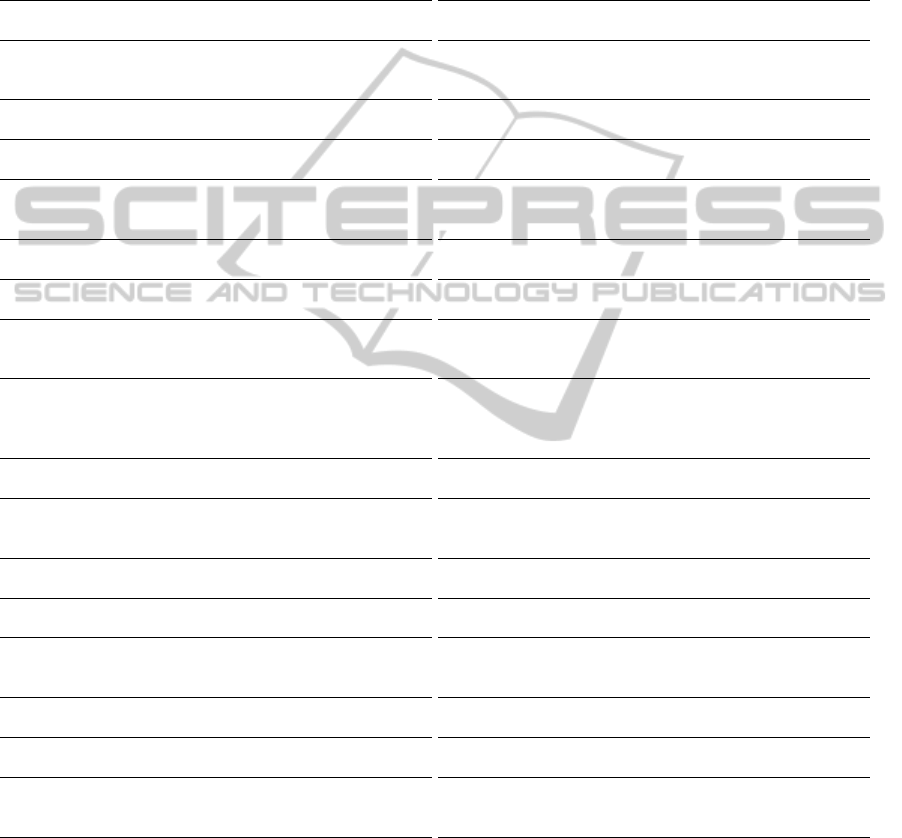
Table 1: A comparison between the Adapted Na
¨
ıve Bayes classifier (ANB), the multinomial Na
¨
ıve Bayes classifier trained
on all source data (MNB s), the multinomial Na
¨
ıve Bayes classifier trained on 5% target data (MNB 5t), and the multinomial
Na
¨
ıve Bayes classifier trained on 80% target data (MNB 80t). The results are reported as average auROC values over five-
fold cross validation trials. For the ANB classifier, the row headings indicated how much target unlabeled data was used in
training the classifier and the column headings indicate how much target labeled data was used. The best values for the ANB
are highlighted. Note that ANB is bounded by MNB 5t and MNB 80t, and that ANB predicts more accurately as the length
of k-mers increases.
PSORTb dataset TargetP dataset
Gram-positive as source and gram-negative as target Plant as source and non-plant as target
Features are 1-mers Features are 1-mers
ANB
MNB s MNB 5t MNB 80t
ANB
MNB s MNB 5t MNB 80t
tTU\tTL 5% 10% 20% tTU\tTL 5% 10% 20%
20% 0.9142 0.9170 0.9208
0.9274 0.9218 0.9352
20% 0.6526 0.6984 0.7398
0.7638 0.7990 0.812840% 0.9068 0.9082 0.9168 40% 0.6290 0.6624 0.6916
60% 0.8900 0.9020 0.9190 60% 0.6088 0.6452 0.7040
Features are 2-mers Features are 2-mers
ANB
MNB s MNB 5t MNB 80t
ANB
MNB s MNB 5t MNB 80t
tTU\tTL 5% 10% 20% tTU\tTL 5% 10% 20%
20% 0.9358 0.9366 0.9394
0.9330 0.9190 0.9424
20% 0.6578 0.7184 0.7938
0.7862 0.8260 0.839640% 0.9284 0.9268 0.9390 40% 0.6212 0.6702 0.6934
60% 0.9292 0.9358 0.9350 60% 0.6028 0.6308 0.6714
Features are 3-mers Features are 3-mers
ANB
MNB s MNB 5t MNB 80t
ANB
MNB s MNB 5t MNB 80t
tTU\tTL 5% 10% 20% tTU\tTL 5% 10% 20%
20% 0.9380 0.9380 0.9424
0.9194 0.8580 0.9552
20% 0.7582 0.8144 0.8566
0.6682 0.6386 0.883640% 0.9262 0.9278 0.9314 40% 0.7404 0.7972 0.8346
60% 0.9134 0.9240 0.9308 60% 0.7618 0.7636 0.7796
Gram-negative as source and gram-positive as target Non-plant as source and plant as target
Features are 1-mers Features are 1-mers
ANB
MNB s MNB 5t MNB 80t
ANB
MNB s MNB 5t MNB 80t
tTU\tTL 5% 10% 20% tTU\tTL 5% 10% 20%
20% 0.9278 0.9320 0.9346
0.9360 0.9142 0.9556
20% 0.7296 0.7190 0.7704
0.7618 0.7366 0.851440% 0.8978 0.9326 0.9118 40% 0.6922 0.7196 0.7696
60% 0.8912 0.8728 0.9302 60% 0.6716 0.7340 0.7548
Features are 2-mers Features are 2-mers
ANB
MNB s MNB 5t MNB 80t
ANB
MNB s MNB 5t MNB 80t
tTU\tTL 5% 10% 20% tTU\tTL 5% 10% 20%
20% 0.9090 0.9452 0.9466
0.9442 0.8852 0.9616
20% 0.7824 0.7810 0.7868
0.7836 0.7508 0.885240% 0.9180 0.9206 0.9502 40% 0.7272 0.7514 0.7862
60% 0.9426 0.9428 0.9428 60% 0.7380 0.7362 0.7592
Features are 3-mers Features are 3-mers
ANB
MNB s MNB 5t MNB 80t
ANB
MNB s MNB 5t MNB 80t
tTU\tTL 5% 10% 20% tTU\tTL 5% 10% 20%
20% 0.9590 0.9520 0.9614
0.9578 0.8118 0.9544
20% 0.8200 0.8092 0.8596
0.8968 0.6860 0.862840% 0.9280 0.9440 0.9460 40% 0.7382 0.7442 0.7990
60% 0.9278 0.9282 0.9460 60% 0.6904 0.7256 0.7848
This dataset contains DNA sequences of 141 base
pairs centered around the donor splice site dimer AG
and the label of whether or not that AG dimer is a
true splice site. The sequences are from five organ-
isms, C.elegans as the source domain, and C.remanei,
P.pacificus, D.melanogaster, and A.thaliana as target
domains. We used the dataset with 100,000 instances
for the source domain, and the datasets with 2,500,
6,500, 16,000, and 40,000 instances for the target do-
main. In each dataset there are about 1% positive
instances. Accurately predicting splice sites is im-
portant for genome annotation (R
¨
atsch et al., 2007;
Bernal et al., 2007).
For this dataset we used the area under precision-
recall curve (auPRC), a metric that is preferred over
area under a receiver operating characteristic curve
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
68

when the class distribution is skewed, which is the
case with this dataset.
The results for this dataset were very poor, with
our algorithm always gravitating towards classifying
each instance as not containing a splice site. We be-
lieve that this is due mainly because the k-mers indi-
cating a splice site occur with low frequency and their
relative position to the splice site is important. We
will discuss in Section 4 how we propose to address
this issue in future work.
4 CONCLUSIONS AND FUTURE
WORK
In this paper, we proposed a domain adaptation classi-
fier for biological sequences. This algorithm showed
promising classification performance in our experi-
ments. Our analysis indicates that the closer the tar-
get domain is to the source domain the better is the
classifier learned. Other conclusions drawn from our
observations: using 2-mers or 3-mers results in bet-
ter prediction, with small differences between them;
removing features from the target domain reduces the
accuracy of classifier; having more target labeled data
increases the accuracy of classifier; and adding too
much target unlabeled data decreases the accuracy of
classifier.
In future work we would like to investigate how
would assigning different weights to the data used for
training influence the accuracy prediction of the algo-
rithm. We would like to assign higher weight to the
labeled data from the target domain since this is more
likely to correctly predict the class of the target test
data than the labeled data from the source domain or
the unlabeled data from the target domain.
We would also like to evaluate other methods for
selecting the generalizable features. For example,
we would like to investigate if selecting generalizable
features using the mutual information of the features
instead of their probabilities, in Equation (1), leads to
better classification accuracy.
Another aspect we would like to improve is the
accuracy of this classifier on the splice site dataset, to
get accuracy that is similar to state of the art splice
site classifiers, e.g., SVM classifiers. We would like
to reduce the number of motifs with different cluster-
ing strategies, and identify more discriminative mo-
tifs using Gibbs sampling or MEME. In addition, we
would like to run experiments on smaller splice site
datasets to better understand the characteristics of this
problem.
ACKNOWLEDGEMENTS
The computing for this project was performed on the
Beocat Research Cluster at Kansas State University,
which is funded in part by NSF grants CNS-1006860,
EPS-1006860, EPS-0919443, and MRI-1126709.
REFERENCES
Baten, A., Chang, B., Halgamuge, S., and Li, J. (2006).
Splice site identification using probabilistic param-
eters and svm classification. BMC Bioinformatics,
7(Suppl 5):S15.
Bernal, A., Crammer, K., Hatzigeorgiou, A., and Pereira,
F. (2007). Global discriminative learning for higher-
accuracy computational gene prediction. PLoS Com-
put Biol, 3(3):e54.
Brown, M. P. S., Grundy, W. N., Lin, D., Cristianini, N.,
Sugnet, C., Furey, T. S., M.Ares, J., and Haussler, D.
(2000). Knowledge-based analysis of microarray gene
expression data using support vector machines. PNAS,
97(1):262–267.
Dai, W., Xue, G., Yang, Q., and Yu, Y. (2007). Transferring
na
¨
ıve bayes classifiers for text classification. In Pro-
ceedings of the 22nd AAAI Conference on Artificial
Intelligence.
Degroeve, S., Saeys, Y., De Baets, B., Rouz
´
e, P., and Van
De Peer, Y. (2005). Splicemachine: predicting splice
sites from high-dimensional local context representa-
tions. Bioinformatics, 21(8):1332–1338.
Eaton, J. W., Bateman, D., and Hauberg, S. (2008). GNU
Octave Manual Version 3. Network Theory Ltd.
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne,
G. (2000). Predicting subcellular localization of pro-
teins based on their N-terminal amino acid sequence.
Journal of molecular biology, 300(4):1005–1016.
Gardy, J. L., Laird, M. R., Chen, F., Rey, S., Walsh, C. J.,
Ester, M., and Brinkman, F. S. L. (2005). Psortb
v.2.0: Expanded prediction of bacterial protein sub-
cellular localization and insights gained from compar-
ative proteome analysis. Bioinformatics, 21(5):617–
623.
Gardy, J. L., Spencer, C., Wang, K., Ester, M., Tusn
´
ady,
G. E., Simon, I., Hua, S., deFays, K., Lambert, C.,
Nakai, K., and Brinkman, F. S. (2003). Psort-b:
improving protein subcellular localization prediction
for gram-negative bacteria. Nucleic Acids Research,
31(13):3613–3617.
Huang, J., Li, T., Chen, K., and Wu, J. (2006). An approach
of encoding for prediction of splice sites using svm.
Biochimie, 88:923–9.
Jaakkola, T. S. and Haussler, D. (1999). Exploiting gen-
erative models in discriminative classifiers. In Pro-
ceedings of the 1998 conference on Advances in neu-
ral information processing systems II, pages 487–493,
Cambridge, MA, USA. MIT Press.
Jiang, J. and Zhai, C. (2007). A two-stage approach to do-
main adaptation for statistical classifiers. In Proceed-
ings of the sixteenth ACM conference on Conference
NaïveBayesDomainAdaptationforBiologicalSequences
69

on information and knowledge management, CIKM
’07, pages 401–410, New York, NY, USA. ACM.
Lorena, A. C. and de Carvalho, A. C. P. L. F. (2003).
Human splice site identification with multiclass sup-
port vector machines and bagging. In Proceedings of
the 2003 joint international conference on Artificial
neural networks and neural information processing,
ICANN/ICONIP’03, pages 234–241, Berlin, Heidel-
berg. Springer-Verlag.
Maeireizo, B., Litman, D., and Hwa, R. (2004). Co-training
for predicting emotions with spoken dialogue data. In
Proceedings of the ACL 2004 on Interactive poster
and demonstration sessions, ACLdemo ’04, Strouds-
burg, PA, USA. Association for Computational Lin-
guistics.
Mccallum, A. and Nigam, K. (1998). A comparison of event
models for naive bayes text classification. In AAAI-98
Workshop on ’Learning for Text Categorization’.
M
¨
uller, K.-R., Mika, S., R
¨
atsch, G., Tsuda, S., and
Sch
¨
olkopf, B. (2001). An introduction to kernel-based
learning algorithms. IEEE Transactions on Neural
Networks, 12(2):181–202.
Nigam, K., Mccallum, A., Thrun, S., and Mitchell, T.
(1999). Text classification from labeled and unlabeled
documents using EM. In Machine Learning, pages
103–134.
Noble, W. S. (2006). What is a support vector machine?
Nat Biotech, 24(12):1565–1567.
Pan, S. J. and Yang, Q. (2010). A survey on transfer learn-
ing. IEEE Transactions on Knowledge and Data En-
gineering, 22(10):1345–1359.
R
¨
atsch, G. and Sonnenburg, S. (2004). Accurate splice site
detection for caenorhabditis elegans. In B. Schlkopf,
K. T. and Vert, J.-P., editors, Kernel Methods in Com-
putational Biology, pages 277–298. MIT Press.
R
¨
atsch, G., Sonnenburg, S., Srinivasan, J., Witte, H.,
M
¨
uller, K.-R., Sommer, R., and Sch
¨
olkopf, B. (2007).
Improving the c. elegans genome annotation using
machine learning. PLoS Computational Biology,
3:e20.
Riloff, E., Wiebe, J., and Wilson, T. (2003). Learning sub-
jective nouns using extraction pattern bootstrapping.
In Proceedings of the seventh conference on Natural
language learning at HLT-NAACL 2003 - Volume 4,
CONLL ’03, pages 25–32, Stroudsburg, PA, USA.
Association for Computational Linguistics.
Sch
¨
olkopf, B. and Smola, A. J. (2001). Learning with Ker-
nels: Support Vector Machines, Regularization, Opti-
mization, and Beyond. MIT Press, Cambridge, MA,
USA.
Schweikert, G., Widmer, C., Sch
¨
olkopf, B., and R
¨
atsch,
G. (2008). An empirical analysis of domain adap-
tation algorithms for genomic sequence analysis. In
NIPS’08, pages 1433–1440.
Sonnenburg, S., R
¨
atsch, G., Jagota, A., and M
¨
uller, K.-R.
(2002). New methods for splice-site recognition. In
In Proceedings of the International Conference on Ar-
tifical Neural Networks., pages 329–336. Copyright
by Springer.
Sonnenburg, S., Schweikert, G., Philips, P., Behr, J., and
R
¨
atsch, G. (2007). Accurate splice site prediction us-
ing support vector machines. BMC Bioinformatics,
8(Supplement 10):1–16.
Tan, S., Cheng, X., Wang, Y., and Xu, H. (2009). Adapting
naive bayes to domain adaptation for sentiment anal-
ysis. In Proceedings of the 31th European Confer-
ence on IR Research on Advances in Information Re-
trieval, ECIR ’09, pages 337–349, Berlin, Heidelberg.
Springer-Verlag.
Tsuda, K., Kawanabe, M., R
¨
atsch, G., Sonnenburg, S.,
and M
¨
uller, K.-R. (2002). A new discriminative
kernel from probabilistic models. Neural Comput.,
14(10):2397–2414.
Vapnik, V. N. (1995). The nature of statistical learning the-
ory. Springer-Verlag New York, Inc., New York, NY,
USA.
Yarowsky, D. (1995). Unsupervised word sense disam-
biguation rivaling supervised methods. In Proceed-
ings of the 33rd annual meeting on Association for
Computational Linguistics, ACL ’95, pages 189–196,
Stroudsburg, PA, USA. Association for Computa-
tional Linguistics.
Zhang, Y., Chu, C.-H., Chen, Y., Zha, H., and Ji, X. (2006).
Splice site prediction using support vector machines
with a bayes kernel. Expert Syst. Appl., 30(1):73–81.
Zien, A., R
¨
atsch, G., Mika, S., Sch
¨
olkopf, B., Lengauer, T.,
and M
¨
uller, K.-R. (2000). Engineering support vec-
tor machine kernels that recognize translation initia-
tion sites. Bioinformatics, 16(9):799–807.
BIOINFORMATICS2013-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
70
