
Eigen Heartbeats for User Identification
Marta S. Santos
1
, Ana L. Fred
1,2
, Hugo Silva
1,2
and André Lourenço
2,3
1
Intituto Superior Técnico, Lisboa, Portugal
2
Instituto de Telecomunicações, Lisboa, Portugal
3
Instituto Superior de Engenharia de Lisboa, Lisboa, Portugal
Keywords: ECG, Principal Components Analysis, User Identification.
Abstract: Electrocardiographic (ECG) signals record the heart’s electrical activity over time. These signals have
typically been assessed for clinical purposes providing a fair evaluation of the heart’s condition. However, it
has been shown recently that they also convey distinctive information that can be used for user
identification. In this paper we explore these signals for user identification purposes, proposing two data
representation and processing techniques based on principal component analysis (PCA) and classification
based on the K-NN rule. We analyze and compare these techniques, showing experimentally that 100%
identification rates can be achieved. The analysis covers an outlier removal procedure and different
configurations of algorithmic and proposed system parameters.
1 INTRODUCTION
In this paper, we analyze the heartbeat as a biometric
modality. The heartbeat is a complex electric signal
produced by the heart; among other advantages, it is
difficult to forge and provides intrinsic aliveness
detection. Targeting increased usability and minimal
intrusiveness, we used a one-lead ECG acquisition
setup with dry electrodes (Lourenço et al., 2011).
Increased noise is inherent to this configuration
requiring adequate pre-processing.
There are two main approaches for template
representation: fiducial, and non-fiducial. The first
consists of finding points of interest within the
segmented ECG (fiducial points), and then
representing heartbeat waves as a set of extracted
features (Biel et al., 2001), (Silva et al., 2007). Non-
fiducial methods, on the other hand, consider the
signal’s shape as the templates (Chan et al., 2008).
We propose a partially fiducial approach. For the
segmentation of the ECG signal into heartbeats we
use a fiducial method, in which the R peak is
detected and used as alignment reference. Then, a
non-fiducial method, based on PCA, is applied for
representation of the heartbeats either describing a
prototypical heartbeat pattern for the overall
population as a global eigen-heartbeat, or expressing
prototypical patterns for each individual. Our work is
closely related with the proposals by (Irvine et al.,
2009) and (Israel et al., 2010), where PCA is also
used. However, while in previous works an eigen-
heartbeat is computed from the overall population,
we propose individualized eigen-heartbeats
computations to characterize each subject. This
methodology has the advantage of easier upgrade of
databases, not requiring the re-computation of new
base eigen-heartbeats as new users are included into
the database. Furthermore, we include an outlier
removal step in our pre-processing phase, aiming the
achievement of better templates and higher
recognition rates. Our work also stands out for using
a larger database than what is found in prior studies,
and ECG acquired at finger/palms.
2 USER IDENTIFICATION
Biometric identification systems typically comprise
two main phases (Jain et al., 2008): enrollment and
identification. At the enrollment phase, the user
provides both his / her identity (e.g. name) and the
biometric template (in our case, the ECG signal).
Afterwards, one or more templates of the acquired
modality are stored in a database for future reference.
The processing phase usually comprises a quality
evaluation procedure; if the acquired signals fail to
accomplish the quality check there is a failure to
enroll (FTE) error.
351
Santos M., Fred A., Silva H. and Lourenço A..
Eigen Heartbeats for User Identification.
DOI: 10.5220/0004249503510355
In Proceedings of the International Conference on Bio-inspired Systems and Signal Processing (BIOSIGNALS-2013), pages 351-355
ISBN: 978-989-8565-36-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
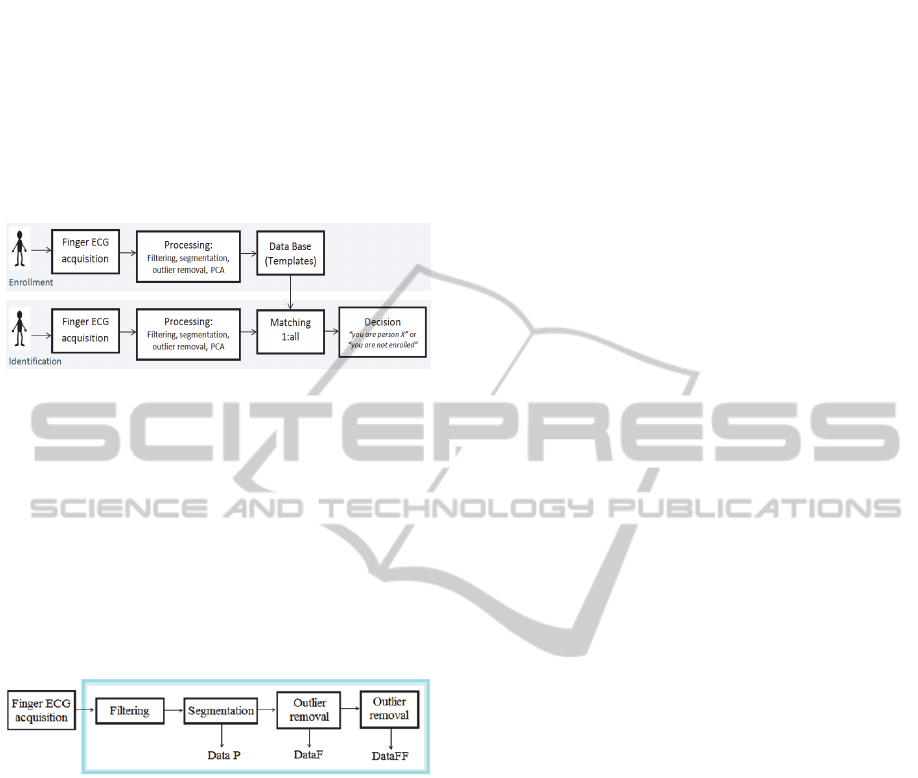
At the identification phase, the user only needs to
present the biometric modality at the sensor level; the
system then validates this data against the templates
previously stored in the database. If the acquired
biometric data shows a match to the template of one
of the enrolled users, an identity will be recognized.
Figure 1 gives a schematic description of the
enrollment and identification phases for the proposed
ECG-based user identification system.
Figure 1: Enrolment and identification stages.
3 PROPOSED METHOD
3.1 Pre-Processing
To deal with the noise inherent to finger/palm
acquisition, we propose a sequence of pre-processing
steps, summarized in Figure 2. Each step is applied
both in the enrollment and identification stages (see
Figure 1) and is described next.
Figure 2: ECG pre-processing steps (framed).
3.1.1 Filtering and Segmentation
The ECG signals measured at hands or palms can be
affected by multiple noise sources. We designed a
FIR band-pass filter (order 301, using a Hamming
window), with 5-20Hz cutoff frequencies. For
segmentation we built on the work by (Engelse et al.,
1979) for offline QRS detection, adapting their
algorithm for real-time operation. Details of the
algorithm and comparison with offline approaches
can be found in (Lourenço et al., 2012).
3.1.2 Outlier Removal
After segmentation, there was a clear need to remove
outliers from the signal (due to excessive noise or
segmentation errors). For that purpose, we created a
simple outlier removal heuristic. Considering the
expected stability of P-QRS complex, we compute
amplitude statistics around certain fiducial points,
considering as outliers segments that present
considerable deviations from the median statistics.
The points chosen were: time instant 75, P wave
neighborhood; 150, just before the Q wave, where
most of the signals show a small amplitude; 200,
where the segments are centered; and 300, just after
the end of the repolarization.
For the ECG recording of each individual, after
signal segmentation, the median of signal amplitudes
at each of the above fiducial points is computed; if, at
least at one of the fiducial points, the absolute
difference between the segments’ amplitude value at
the point and the median is larger than the median
value multiplied by a factor α, the segment is
eliminated; the tuning of this parameter value is
described in the experimental results section. The
outlier removal procedure is applied once or twice in
sequence; the corresponding remaining (clean) data
after the one or two step approach is hereafter
referred as DataF and DataFF, respectively.
3.2 PCA-based Feature Extraction
By applying the PCA technique, each ECG segment
is described as a linear combination of eigen-vectors,
herein referred as eigen-heartbeats. By storing these
eigen-heartbeats and the mean wave, each ECG
segment is described by the coefficients in the linear
combination, forming the set of features that
describes it. The PCA was applied: (a) to the data of
the whole population, leading to an eigen
representation of the average ECG segments pattern;
(b) individually to each person’s ECG segments.
The first method, hereafter referred as the Overall
Population Eigen-Heartbeat (OEigHB) approach,
leads to a database formed by the mean and
eigenvectors corresponding to the whole population
data. Each enrolling ID will be represented by the
coefficients resulting from the projection of their HB
segments into the database eigenvectors and stored as
template for that individual. The second method,
hereafter referred as the Individualized Eigen-
Heartbeat (IEigHB) approach, will have, for each
individual, as template, the mean ECG segment,
eigenvectors and coefficients referent to each
individual, which will be stored in the database.
3.3 Decision
User identification is performed based on the
comparison of the presented biometric modality with
the stored templates for each individual. The
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
352
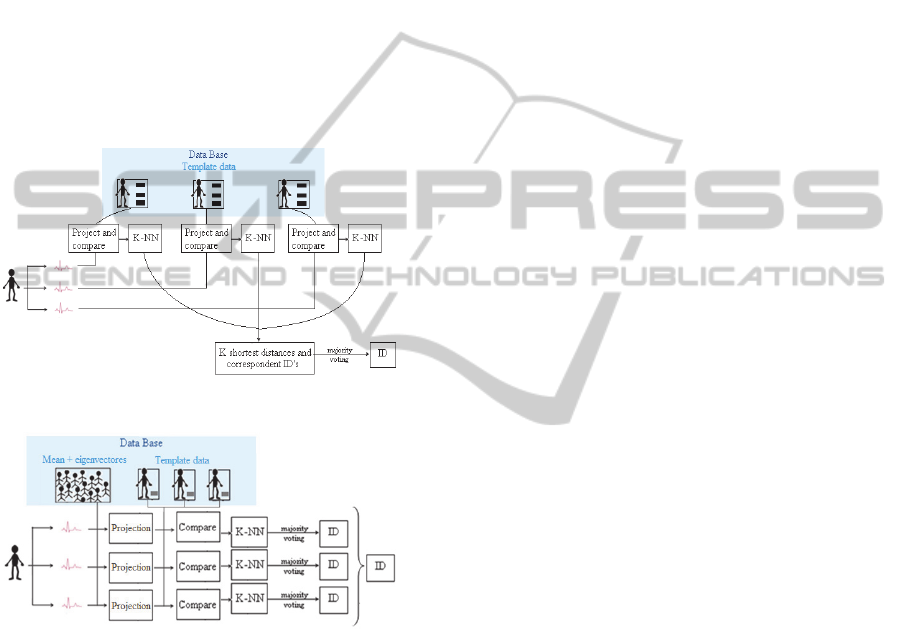
proposed decision algorithm is different for the two
approaches. In the OEigHB identification process,
the newly presented ECG segments (after pre-
processing) are projected into the eigenvectors taken
from the whole population (see Figure 4), and the
obtained feature vectors are matched with the
templates of each enrolled user. According to the
IEigHB approach, the new ECG segments are
projected into each enrolled user’s eigen-heartbeats,
and these projection vectors are compared with the
several users’ templates (see Figure 3). We use
template matching with the Euclidean distance, and
the K-NN algorithm to correspond each new segment
with a user ID. The identity of a given segment is
decided using majority voting, choosing the most
represented identities among the K-NNs templates.
Figure 3: IEigHB identification scheme.
Figure 4: OEigHB identification scheme.
4 EXPERIMENTAL RESULTS
4.1 Datasets
The ECG datasets used in our experimental
evaluation were provided by the Check Your
Biosignals Here initiative (Silva et al., 2011). The
dataset comprises a single session of acquisition of
different physiological signals, including the ECG,
over a population of 65 volunteering individuals, in a
physically unconstrained setup. The experiment
consisted of two distinct moments: an introductory
phase, during which a staff member would explain
the goal and details of the experiment; and an
emotion inducing phase triggered by the visualization
a video sequence. The acquisition time was thus
variable, depending on the duration of the interaction
between the user and the staff member that described
the test. ECG signals were collected from fingers and
palms using dry Ag/AgCl electrodes. It is worth
noting that the video sequence presented triggered
different emotions, which are a source of variability.
4.2 Outlier Removal Procedure
Due to variable acquisition times, the total number of
segments varies for different individuals. Also, there
are a different number of outliers for each individual.
As a result, the α-value, that controls the level of
outlier removal, will lead to different number of
segments kept per user. To standardize the number of
segments used as templates per individual (NS), this
parameter was made constant. Two values were
tested: 20 and 30. In order to evaluate the error
probability of the identification system, we used
independent training and test sets, randomly chosen
from the available data. By training set we denote the
segments used as templates; the remaining segments
per individual, after outlier removal, were used to
assess the identification accuracy (test data).
Error estimates were performed by averaging
over 25 runs of the classification procedure using
randomly selected training and test sets. However,
for some individuals and α-values combinations, the
minimum NS established was not reached. Those
were counted as individuals that “failed to enroll”
(FTE). The value of α will also influence the
identification rates. The lower the value of α, the
higher the number of segments that are kept, leading
to a smaller FTE rates; however, with the
corresponding larger amount of outliers left in the
data with these lower α values, lower identification
error rates are obtained. Considering the two
identification methods, IEigHB and OEigHB, the
lowest identification error rates, with acceptable FTE
rates, were obtained using the outlier removal
procedure with α = 0,4 (which was ultimately,
chosen, and used hereafter in our studies). However,
for large values of NS and k, it is possible to obtain
similar identification errors with lower FTE errors.
4.3 Segments for Enrollment (NS)
We evaluated the influence of the number of
templates per user used on the identification accuracy
of the system. The next plot shows the average and
standard deviation (SD) of the identification error
EigenHeartbeatsforUserIdentification
353
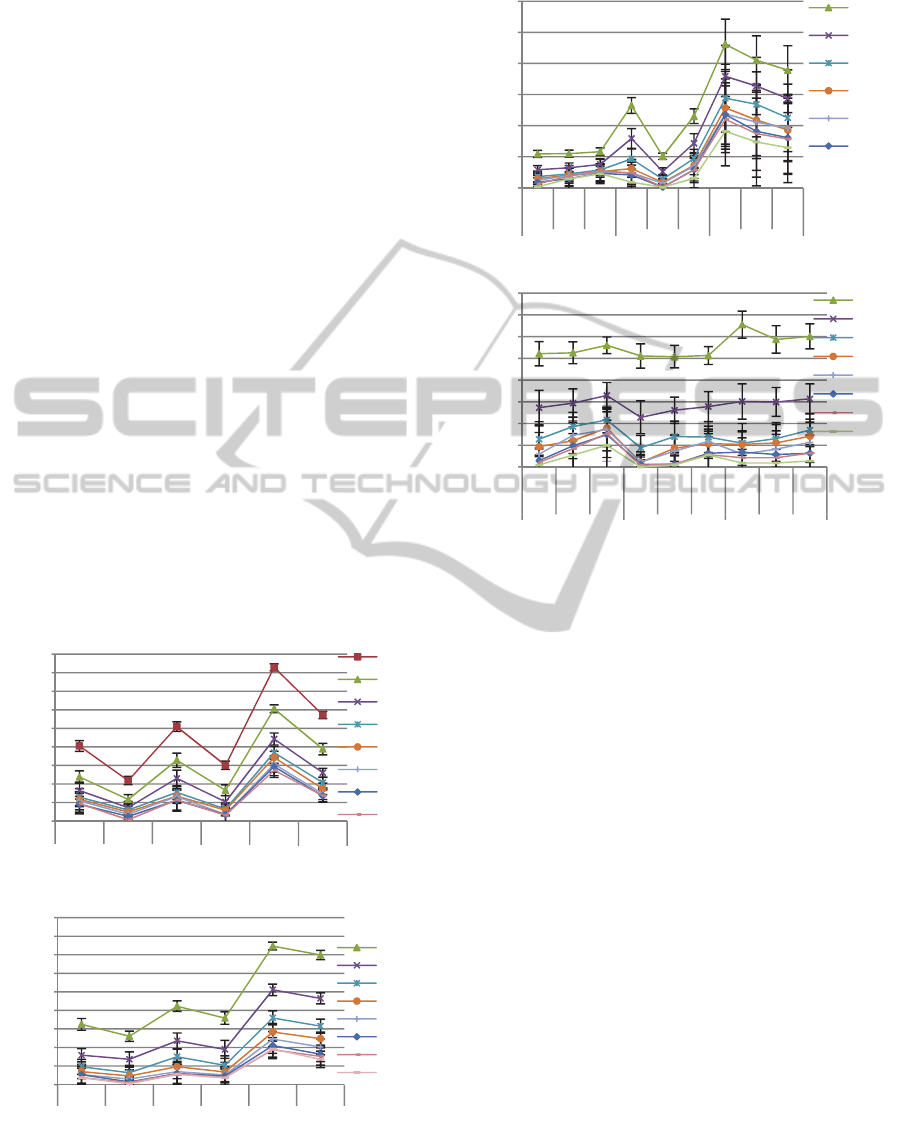
rates using both methods and obtained over 25 runs
using a 1-NN classifier and different values for k, the
number of ECG segments presented for accessing the
system. We tested the situations of NS=20 and
NS=30 for the three setups of: no outlier removal;
outlier removal in one step; and using twice the
outlier removal procedure.
As shown, the higher the NS the lower the error
probability is, and so, longer enrolment times are
preferable. The OEigHB seems to be less sensitive to
the various configurations, having lower ranges on
error rates than the IEigHB approach. More template
segments capture better the variability of the
subject’s heartbeat so new segments are more likely
to mach correctly the ones of the database.
4.4 Segments for Identification (K)
The number of segments required for accessing the
system is closely related with the acquisition time
when trying to identify a subject in a post-enrolment
phase. It is the number of heartbeats that the user
provides to the system in order to be identified.
From Figure 5, for this parameter the larger the
amount of segments, k, the lower the error
probability. However, values of k between 9 and 15
lead to similar error probability. For large k and NS it
can be observed a 0% error probability.
(a)
(b)
Figure 5: (a) IEigHB approach; (b) OEigHB approach. 1-
NN. Each line represents the k parameter. In the x-axis the
NS parameter and the data used are represented.
(a)
(b)
Figure 6: Error probability variation in (a) IEigHB and (b)
OEigHB identification methods, obtained with dataFF and
NS=30, for different energies and K-NN values.
4.5 Outlier Sensibility
From Figure 5 it is apparent that the identification
error decreases with the decrease of the number of
outliers among the data (compare results with DataP
vs DataF). However, the gain is not so impressive for
the second time one applies the procedure – DataFF
results. Both methods (OEigHB and IEigHB) lead to
error probabilities lower than 5% without outlier
removal (DataP). However, with DataFF both
methods can achieve 0% error rate with small SD.
4.6 Energy
The reduction of the data energy is associated with
the data compression that is obtained by eliminating
some eigenvectors that represent weakly the data
variance. In this dataset, a 5% energy reduction
corresponds approximately, to a 50% data reduction,
and a 10% energy reduction to a 60% data reduction.
Figure 6 plots identification errors for different
scenarios with different compression levels. The
OEigHB method is less prone to error coming from
the data compression, keeping both the mean error
and SD similar with the decrease of the data energy.
0
0,025
0,05
0,075
0,1
0,125
0,15
0,175
0,2
0,225
20 30 20 30 20 30
Data filtered Data filtered Data without
Error probability
IEigenHB KNN =1
1
3
5
7
9
11
13
15
0
0,02
0,04
0,06
0,08
0,1
0,12
0,14
0,16
0,18
20 30 20 30 20 30
Data filtered Data filtered Data without
Error probability
OEigenHB KNN =1
1
3
5
7
9
11
13
15
0
0,05
0,1
0,15
0,2
0,25
0,3
KNN=
1
KNN=
3
KNN=
5
KNN=
1
KNN=
3
KNN=
5
KNN=
1
KNN=
3
KNN=
5
Error probability
Individualized Eigen-HB
1
3
5
7
9
1
1
0
0,01
0,02
0,03
0,04
0,05
0,06
0,07
0,08
KNN=1
KNN=3
KNN=5
KNN=1
KNN=3
KNN=5
KNN=1
KNN=3
KNN=5
Error probability
Population Eigen-HB
1
3
5
7
9
11
13
15
DataP
DataF
DataFF
DataP
DataF DataFF
BIOSIGNALS2013-InternationalConferenceonBio-inspiredSystemsandSignalProcessing
354

On the other hand, for the IEigHB method, both the
mean and SD increase with lower energy values.
4.7 K- Nearest Neighbours (K-NN)
As depicted in Figure 6, the number of K-Nearest
Neighbors used in the decision method does not seem
to influence significantly the error probability’s
mean. However, there is a general tendency that for
lower energys and low values of k, the better results
are obtained by increasing the KNN. This indicates
that when the data is poorer (less segments, less
energy) it is advisable to use a higher number of
neighbours in the decision process in order to lower
the identification error.
5 CONCLUSION
In this paper we presented a framework and
methodology for user identification from ECG
signals exploring PCA and K-NN classification,
combined with outlier removal. Two main
approaches were proposed, either using eigen-
heartbeats that model the overall population, or using
individualized eigen-heartbeats per user. Overall,
both methods have the potential of successfully
identifying individuals using their finger/hands ECG
signal. Using 30 heartbeat segments as templates,
and 10 segments for accessing the system, both
methods lead to a 0% identification error. However,
the OEigHB method has shown, in general, lower
sensitivity to the design parameters, presenting the
lowest error values. Emotional and pathological
states may create intra subject variability through
time, lowering the accuracies obtained. This topic
has not been thoroughly studied in the context of
biometrics; however some work is already trying to
account for these factors (Agrafioti, 2011).
The down side of ECG-based biometric methods
is that longer enrolment and identification time is
needed to achieve better accuracies (around 30 sec.
of enrollment and 10 sec. of access time for a 0%
error). However, this biometric modality is less
prone to forging than more conventional modalities,
such as the fingerprint, and can verify aliveness and
stress level, which can be useful to prevent
unwillingly identification. Ongoing work includes a
further enlargement of the database, and extending
this study to situations of multiple acquisitions at
distinct time instants.
ACKNOWLEDGEMENTS
This work was partially funded by Fundação para a
Ciência e Tecnologia (FCT) under grants SFRH/
BD/65248/2009 and SFRH/PROTEC/49512/2009.
REFERENCES
Agrafioti, Foteini (2011). PhD thesis. Univ. of Toronto.
Biel, L.; Pettersson, O.; Philipson, L. and Wide, P. (2001)
ECG analysis – a new approach in human
identification. IEEE Transactions on Instrumentation
and Measurement, 50(3):808–812.
Chan, A.; Hamdy, M.; Badre, A. and Badee, V. (2008).
Wavelet distance measure for person identification
using electrocardiograms. IEEE Transactions on
Instrumentation and Measurement, 57(2):248–253,
Engelse, W. A. H.; and Zeelenberg, C. (1979). “A single
scan algorithm for QRS-detection and feature
extraction,” Comp.in Card., vol. 6, pp. 37–42.
Irvine, J.; A., S. (2009). eigenPulse: Robust Human
Identification from Cardiovascular Function. The
Draper Technology Digest , pp. 50-59.
Israel, S., M., J. (2010). The Heartbeat: the Living
Biometric. Chapter in Biometrics: theory, Methods and
Applications, pp. 429-459. Wiley
Jain, A. and Flynn, P. and Ross, A. (2008). Handbook of
Biometrics. Springer.
Lourenço, A.; Silva, H.; Fred, A. (2011). Unveiling the
Biometric Potential of Finger-Based ECG Signals.
Computational Intelligence and Neuroscience
Lourenço, A.; Silva, H.; Leite, P.; Lourenco, R., and Fred,
A. (2011). “Real time electrocardiogram segmentation
for finger based ECG biometric,” in Proc. of
BIOSIGNALS 2012, pp. 49–54.
Silva, H., Fred, A. L., and Lourenço, A. (2011). Check
Your Biosignals Here: Experiments on Affective
Computing and Behavioral Biometrics. RecPad.
Silva, H.; Gamboa, H. and Fred, A. (2007). One lead ECG
based personal identification with feature subspace
ensembles. In Proc. of the MLDM ‘07, pp. 770–783.
EigenHeartbeatsforUserIdentification
355
