
Cloud based Services for Biomedical Image Analysis
D. Wang, T. Bednarz, Y. Arzhaeva, P. Szul, S. Chen, N. Burdett, A. Khassapov,
T. Gureyev and J. Taylor
CSIRO Mathematics, Informatics and Statistics, Commonwealth Scientific and Industrial Research Organization (CSIRO)
11 Julius Avenue, North Ryde, Sydney, NSW 2113, Australia
Keywords: Cloud Computing, Image Processing, Image Analysis, Visualization, Workflow, High Performance
Computing.
Abstract: With the software as a service becoming an increasingly prevalent delivery model, we have developed a
cloud-based image analysis toolbox to provide a wider user base with easy access to the software tools that
we have developed over the last decade. This paper discusses our work on the design and implementation of
the cloud-based image analysis and processing services on an Australian national cloud infrastructure,
including their architecture, workflow management framework, image analysis and visualization examples,
and the challenges we faced. Key components of the services are described, showing the capabilities of the
service engine for real-world cloud-based biomedical image analysis applications.
1 INTRODUCTION
Cloud computing is an emerging infrastructure
paradigm that provides more scalable and efficient
solutions to large scale computing tasks. The
adoption of Web browsers as a standard user
interface and increased Internet bandwidth have
fueled the widespread use of cloud computing
(Monaco, 2012; Pasik, 2012). Large-scale
computing and data storage centers have been
implemented to provide cloud computing services
(Amazon, 2012; Microsoft, 2012; Google, 2012).
Cloud computing also holds great promises to
eliminate the need for maintaining expensive
computing facilities while offering a large pool of
easily accessible, virtualized resources, which are
dynamically reconfigurable on demand in a highly
automated fashion.
Cloud-based image analysis applications have
been proposed, implemented and deployed in
various areas by both industry and academia. Smart
Imaging Technologies Co. developed cloud-based
image analysis software to run complex automated
image analysis on composite multi-layered images,
to create data overlays, and to download the image
analysis results from the measurement databases
(SIMAGIS, 2012). Siemens has brought all of its
cloud activities at its Competence Centre so as to be
able to provide cloud computing services to
individual company sectors from a central point
(Siemens, 2012). One of the proposed applications is
remote diagnostics using cloud based medical
image-processing. Hospitals will be able to send
medical images to a Siemens service centre to
process the images and receive the diagnosis results
as a cloud computing service. PCI Geomatics
released a white paper in July 2011 claiming on-
demand satellite image processing would be the next
generation technology for processing terabytes of
imagery on the cloud (PCI, 2011). A workflow
framework for cloud computing is described and
evaluated with image processing applications to
analyze images from the solar system (Shams et al.,
2010). Almeer et al. presented a case study to
process remote sensing images using Hadoop
MapReduce framework in cloud environment
(Almeer, 2012). Cloud computing study has also
been conducted to evaluate an image mosaic engine
and its application to the creation of image mosaics
and management of their provenance (Berriman et
al., 2010). Golpayegani et al. reported their work
using cloud computing for processing satellite data
on high end computer clusters with Hadoop
Distributed File System (HDFS) and MapReduce
framework (Golpayegani and Halem, 2009). More
recently, a real-time face recognition application was
350
Wang D., Bednarz T., Arzhaeva Y., Szul P., Chen S., Burdett N., Khassapov A., Gureyev T. and Taylor J..
Cloud based Services for Biomedical Image Analysis.
DOI: 10.5220/0004370003500357
In Proceedings of the 3rd International Conference on Cloud Computing and Services Science (CLOSER-2013), pages 350-357
ISBN: 978-989-8565-52-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

presented by Soyata et al. (Soyata et al., 2012) using
their mobile-cloudlet-cloud architecture named
MOCHA. Ferzli and Khalife also presented their
mobile cloud computing educational tool for image
and video processing algorithms (Ferzli and Khalife,
2011).
While the above works explored different aspects
of cloud computing on specific platforms and
applications in various domains, this paper presents
our project which is concerned with designing a
novel cloud-based image analysis and processing
toolbox on a national cloud infrastructure, including
its architecture and implementation. The project is
directly inspired and funded by the Australian
Government initiatives of National eResearch
Collaboration Tools and Resources (NeCTAR)
(NeCTAR, 2012). The initiatives are aimed at
building a new infrastructure using existing and new
information and communications technologies.
NeCTAR has four main program areas including
Virtual Laboratories, Research Cloud, eResearch
Tools and The National Servers program. The
research cloud is a highly scalable, cost-effective
and self-service platform, comprising eight
distributed nodes and up to 30,000 CPU cores. Our
cloud-based image analysis toolbox is designed as
eResearch Tools to run on the Research Cloud. It
will be hosted in the Characterization Virtual
Laboratory and the Genomics Virtual Laboratory
which are also part of NeCTAR.
The project focuses on the integrations of various
software components, including the workflow
management framework Galaxy (Galaxy, 2012),
CloudMan (CloudMan, 2012), SGE Job Manager
(Oracle, 2012), various image analysis components,
an interactive image visualization component, an
automated job distribution component for large
image dataset processing, GPU utilization for both
image processing and visualization, and a data
storage management component. The challenges of
the project include how to seamlessly integrate these
components in a cloud infrastructure environment,
how to address the data security and privacy issue,
how to transfer and manage intensive, complex and
big image datasets, and how to monitor and
supervise usage and performance of the cloud based
image analysis services. Our contributions described
in this paper are summarized as follows:
(1) We utilized various frameworks for data
intensive computations and seamlessly integrated
them into a single cloud based service platform
for deploying various applications. To the best of
our knowledge, no prior work has yet shown this
type of results in a large-scale national cloud
infrastructure.
(2) We demonstrate the capabilities of our cloud-
based image analysis and visualization toolbox
using various real-life applications. The toolbox
provides an easy way for various user
communities to access the well-established
image processing and analysis algorithms and
software as services without knowing and caring
details about these algorithms and how and
where they are executed.
The rest of the paper is organized as follows. In
Section II, we describe the architecture of our cloud
based image analysis services, providing
information about each component. Section III
details the workflow management framework used
in the cloud-based image analysis services. The tools
provided in the cloud-based services are described in
Section IV. In Section V, we show the feasibility
and usefulness of our toolbox using a number of
real-world applications for biomedical image
analysis. Finally, we summarize our results and
discuss the future work in Section VI.
2 ARCHITECTURE OF
CLOUD-BASED IMAGE
ANALYSIS SERVICES
The cloud-base image analysis and processing
toolbox comprises a collection of physical and
virtualized resources connected through networks,
including the NeCTAR research cloud Infrastructure
as a Service (IaaS), cloud enabled image analysis
and processing Platform as a Service (PaaS), and our
image analysis Software as a Service (SaaS), which
can be accessed by users through a web portal.
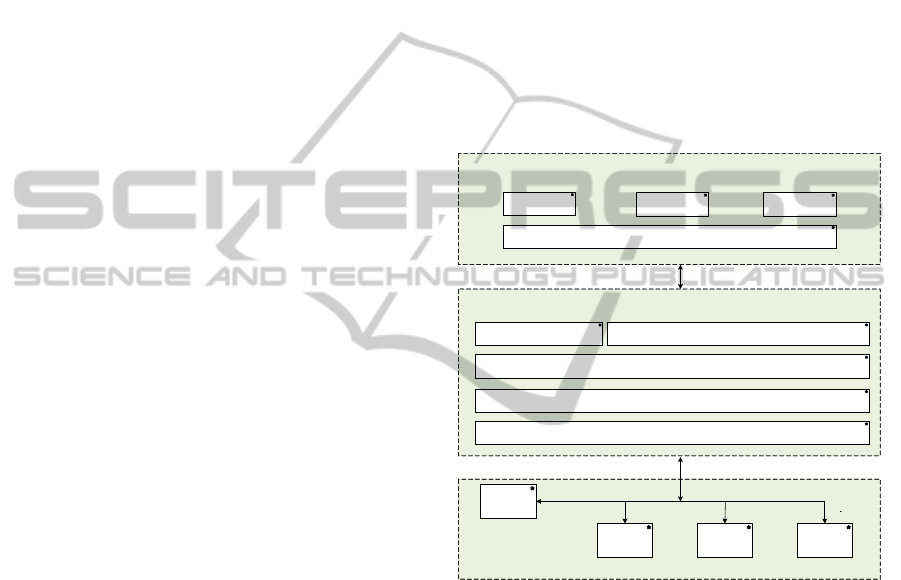
Figure 1 shows a high-level architectural view of the
cloud-based services, including three layers, namely
the NeCTAR research cloud infrastructure layer, the
cloud enabled image analysis and processing
platform layer, and the image analysis and
processing tools layer.
The NeCTAR research cloud delivers basic
compute capabilities as standard services over the
Internet. Servers, storage systems and network
resources are pooled and made available to be
allocated and configured for different applications.
The NeCTAR cloud uses the OpenStack cloud
operating system which is designed to provide
flexibility with no proprietary hardware or software
requirements (OpenStack, 2012). The OpenStack
cloud operating system provides three shared
services: Compute, Networking and Storage. The
CloudbasedServicesforBiomedicalImageAnalysis
351
Compute provisions and manages large networks of
virtual machines; the Networking provides
pluggable, scalable, API-driven network and IP
management services; and the Storage provides
services of object and block storage for use with
servers and applications.
2.1 Platform as a Service
The image analysis and processing platform (PaaS)
represents the development and runtime
environment where the image analysis and
processing tools are executed. The platform also
provides the basic management features of the single
node and leverages all the other operations on the
services that it is hosting. The services include task
submission, job and resource scheduling, error
handling, reporting (traffic, client demands and
usage), execution of the tools, operation status and
progress monitoring, results returning etc. The
platform encapsulates a layer of software and
provides it as a service that can be used to build high
level image analysis and reconstruction services.
The software encapsulated in the platform includes
CloudMan, Galaxy, SGE Job Manager, HTTP, FTP,
SVN, MySQL, Perl, Tcl, PHP, GNU C Library, gcc,
GDCM (DICOM), libPNG, libTIFF, OpenJPEG,
Pthread, Zlib, SZlib, Boost, ITK, VTK, TinyXML,
libsigc++, Glew, WxWidgets, edtProcs etc. With the
platform, virtualization can be implemented by
building a virtual machine image by laying all of the
above software components and tools onto the
OpenStack image.
2.2 Software as a Service
The SaaS layer features three applications offered as
a service on demand. A single instance of the tools
extracted from each of the image analysis packages
runs on the cloud and services multiple end users.
The software packages include HCA-Vision, X-
TRACT and MILXView, as described below:
1) HCA-Vision has been developed for automating
the process of quantifying cell features in
microscopy images. It can reproducibly analyze
complex cell morphologies. The software
provides utilities to measure the morphology of
cells, particular for neurons.
2) X-TRACT implements a large number of
conventional and advanced algorithms for 2D
and 3D X-ray image analysis and simulation. It
provides tools for reconstruction and simulation
of X-ray phase-contrast CT, including phase
retrieval, parallel filtered back projection (FBP),
cone beam Feldkamp Davis Kress (FDK)
algorithms etc.
3) MILXView is a 3D medical imaging analysis
and visualization platform developed by the
biomedical Imaging team at the Australian e-
Health Research Centre (AEHRC). It was
designed and developed to support internal
research efforts, and provide a viable and robust
environment for clinical applications.
MILXView comprises of a core framework that
includes standard imaging functions such as
windowing, histogram inspection, panning,
slicing, zooming, metadata inspection etc, and a
large number of plug-in components that add
visualization, image analysis functions and
complex image processing pipelines.
Figure 1: The architecture of the cloud enabled image
analysis and processing tools.
3 WORKFLOW MANAGEMENT
FRAMEWORK
Using scientific workflow for developing and
executing data processing and analysis pipelines has
gained wide attention over the past decades. A
workflow is one or more pipelines consisting of a
series of functional steps needed to solve a specific
problem. Biomedical image analysis is typically
conducted using multiple functions and proceeds in
a staged fashion with the output of one function used
as an input of another. Many image analysis
functions can be compute-intensive and their
algorithms need to be parallelized to execute in
OpenStack
Dashboard
OpenStack
Dashboard
OpenStack
Dashboard
OpenStack
Dashboard
OpenStackCloudOS
APIs
CloudMan,Galaxy,
SGEJobManager
GDCM(DICOM),libPNG,libTIFF,OpenJPEG,Pthread,Zlib,SZlib,Boost
Apache,HTTP,FTP,SVN,MYSQL,Perl,Tcl,PHP,GNUCLibrary,gcc
MILXViewX‐TRACT
HCA‐Vision
APIs&Tools
ImageAnalysisandProcessingApplications(SaaS)
CloudEnabledImageAnalysisandProcessingPlatform(PaaS)
WebServices(taskSubmission,Status/ProgressMonitoring,ReturningResultsetc.)
ITK,VTK,TinyXML,libsigc++,Glew,WxWidgets,edtProcs
CLOSER2013-3rdInternationalConferenceonCloudComputingandServicesScience
352
cloud. The workflow system provides a flexible
approach to both developing and executing image
analysis applications and makes use of high
performance computing resources in cloud.
We adopt Galaxy as our workflow engine in our
system, which is an open-source, web based
platform for data intensive biomedical research
(Galaxy, 2012). In our case, we excluded
bioinformatics functions and reuse its web portal for
a user to add a list of image analysis and processing
tools. Each of the tools provides a special user
interface to upload image datasets, to specify image
analysis parameters, such as which images to be
uploaded for processing, which processing and
analysis tools to apply for the images, and the
location where the output images and image analysis
results are going to be stored, and to execute the
tool. As the user submits a sequence of tasks with
the output of one task feeding the input of another,
Galaxy automatically records a history log, which is
then presented to the user as a graphical workflow.
The workflow can be edited and submitted for
further executions if needed.
The WMF also transparently handles the storage
of input and output data of the workflows and
simply provides users the links to access the images
and numerical results of their workflows executions,
with an option to relocate them across the cloud data
repositories. These results are provided in the form
that could be visualized by virtual laboratories.
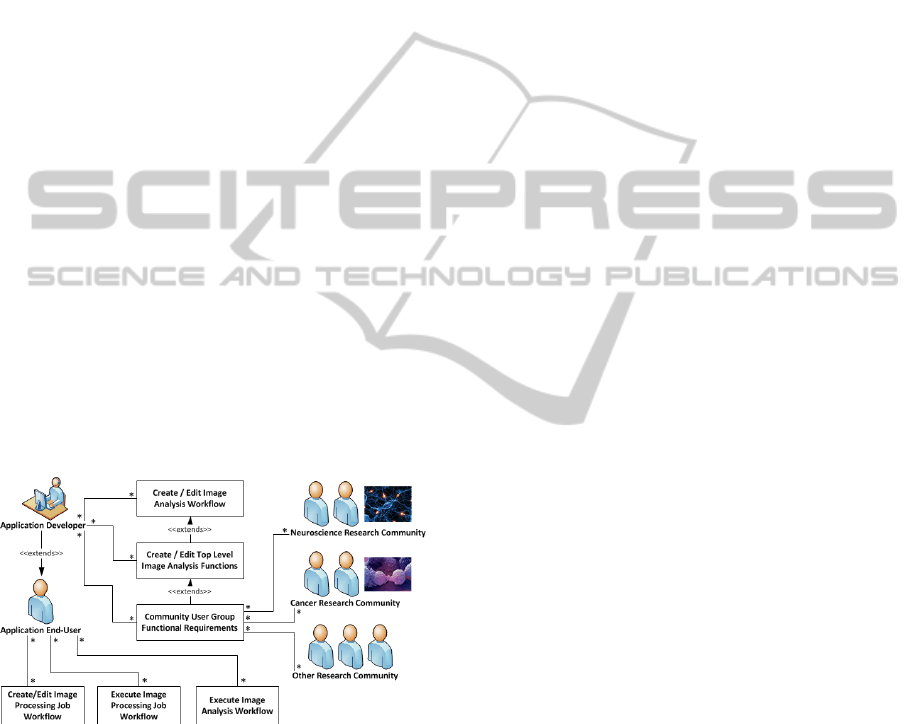
Figure 2: A typical high-level use case for the workflow
environment.
Based on the user community requirements, top-
level image analysis tools can be developed and
included in the toolbox as new services. The
workflows for those commonly used image analysis
routines can be saved for future reuse. The users of
the image analysis services can create their image
analysis jobs as a workflow, and then submit and
execute the workflow via a web portal. A typical
high-level use case for the workflow environment is
shown in Figure 2.
4 CLOUD-BASED IMAGE
ANALYSIS SERVICES
The cloud-based services provide a suite of image
analysis and processing tools. This section describes
some of these tools and shows how they can be used
for image processing and visualization.
4.1 Access NeCTAR Research Cloud
To facilitate end users’ easy access to the NeCTAR
research cloud, it utilizes the Australian Access
Federation Registry (AAF) (AAF, 2012) to provide
a web portal based single sign-on for users with the
same login credentials that they use for login to their
institutional networks.
4.2 Image Analysis Toolboxes
Figure 3 shows the graphical user interface of the
toolbox after login to the cloud system. On the left
hand side, a list of image analysis categories is
displayed, including:
1) Get Data – for a user to upload images, upload
and merge multiple files into a single dataset, or
split a multiple file dataset into standard files.
2) Image Processing – this category contains image
pre-processing tools, including generic
procedures and algorithms that are performed
without a priori knowledge about the specific
features of an image.
3) Image Registration – Included in this category
are tools used to co-register two or more images
in 2D and 3D. The registration is conducted by
transforming different image datasets, acquired
from different modalities or at different time or
different resolutions, into one coordinate system.
The tools for the image registration include
affine transforms, rigid transforms, etc.
4) Image Segmentation – The image segmentation
tools are grouped into this category. It is
composed of different algorithms to divide an
image into connected regions, such as healthy
anatomical structures and pathological tissue.
5) Cellular Image Analysis – The cellular image
analysis tools include automated solutions for
cell image analysis. They include automated
nucleus detection, cytoplasm detection, cell
membrane detection, cell detection, dots and
linear feature detection within a cell, retrieving
CloudbasedServicesforBiomedicalImageAnalysis
353
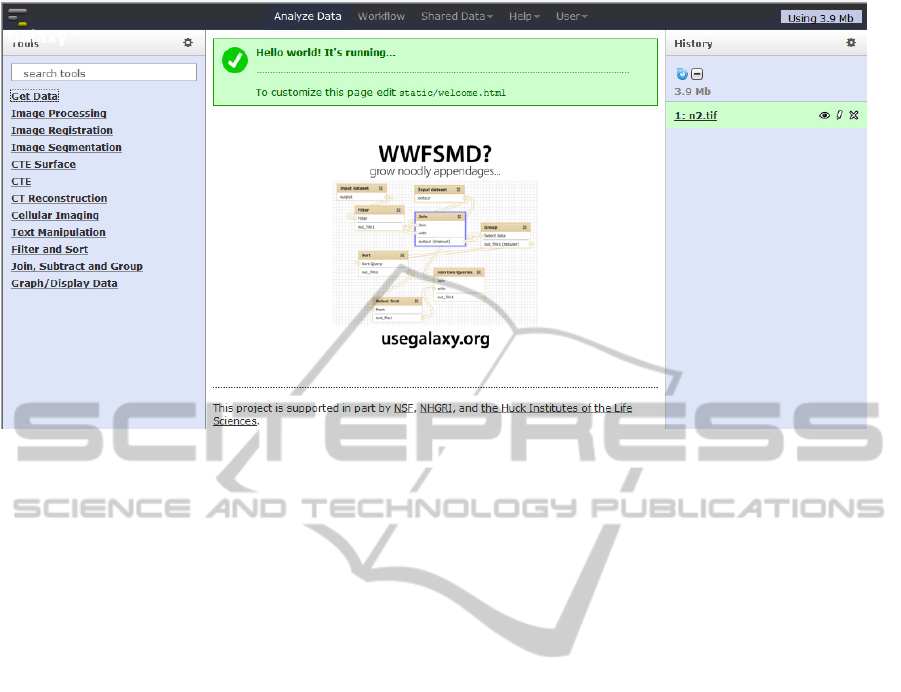
Figure 3: The graphical user interface of the cloud-based image processing and analysis services.
statistical features of dots, lines, membranes,
cytoplasm, cells etc.
6) CT Reconstruction – Tools for CT
Reconstruction include sinogram creation, ring
artifact removal, dark current subtraction, flat
field correction, positional drift correction, data
normalization, Transport of Intensity Equation
(TIE) based phase extraction, Filtered Back-
Projection (FBP) parallel-beam CT
reconstruction, Feldkamp-Davis-Kress (FDK)
cone-beam CT reconstruction, automated
detection of the centre of rotation in a CT scan,
CT reconstruction filters, region of interests
reconstruction, etc.
7) Medical Image Analysis – The medical image
analysis toolbox comprises a suite of functions
for processing and visualizing 3D and 4D
medical images, such as image normalization,
atlas registration, bias field correction, partial
volume estimation, brain topology correction,
cortex thickness estimation, cortical surface
extraction, biomarker mapping on cortical
surface, etc.
5 BIOMEDICAL IMAGE
ANALYSIS APPLICATIONS
This section demonstrates some applications of the
cloud-based image analysis services, including tools
for (1) cellular image analysis; (2) extracting 2D
slices from a 3D image; (3) image registration; and
(4) image visualization.
5.1 Cellular Image Analysis
Cellular image analysis attracts the interests of both
pharmaceutical industry and academia. Researchers
can use the cellular image analysis tools to carry out
high content analysis for their biomedical research.
The image analysis tools provided in the cloud-
based services can help them to conduct automated
measurement of cell morphology and analysis of
cellular responses in individual cells treated with
different chemical compounds. In this example
application, we demonstrate how to use the cloud-
based image analysis tools to detect nuclei,
cytoplasm and cells. The procedure is described as
follows:
1) Upload the image to be analyzed using “Get
Data”.
2) Use “Find Nuclei” tool to detect nuclei by
specifying the input image and various
parameters as shown in Figure 4.
3) Use “Find Cells” tool to detect all cells based on
the nuclei detected in the previous step.
4) Use “Find Cytoplasm” to identify cytoplasm
using the nucleus mask produced in Step 2.
5) Use “Extract Workflow” in History menu to
produce the workflow based on the automatically
recorded history log as shown in Figure 5. View,
edit and share the workflow using Workflow
menu of Galaxy. Figure 6 shows the cellular
image analysis workflow produced.
CLOSER2013-3rdInternationalConferenceonCloudComputingandServicesScience
354

Figure 4: Parameter panel for nucleus detection tool.
Figure 5: Automatically recorded history log.
Figure 6: Image analysis workflow for detecting nuclei, cytoplasm and cells.
Figure 7: GUI of “Extract 2D Slice” image processing tool.
CloudbasedServicesforBiomedicalImageAnalysis
355

5.2 Extracting 2D Slices from a
3D Image
The example shown below demonstrates how to
extract 2D image slices from a 3D MRI image.
Figure 7 shows the Graphical User Interface (GUI)
to select a compressed 3D image. It takes only a few
seconds for the “Extract 2D Slice” tool to retrieve
2D slices from the selected 3D image and insert
them into an html page displayed in Figure 8.
Figure 8: HTML page showing the 2D slices extracted
from the 3D MRI image.
Figure 9: Registration results using affine transformation.
Figure 10: Slice:Drop image viewer used in the cloud-
based image analysis toolbox.
The tool also allows users to download the images.
5.3 Image Registration
Image registration is widely used in healthcare and
medical research. Typical applications of image
registration include combining images of the same
subject from different modalities, aligning temporal
sequences of images to compensate for motion of
the subject between scans at different times.
The image registration tool in our toolbox allows
a user to select two images with one being a fixed
image and the other being a moving image. The tool
employs both rigid and affine methods to transform
the moving image. The former allows images to be
rotated and translated while the latter allows scaling
and shearing as well. Figure 9 shows the registration
result produced by the image registration tool.
5.4 Image Visualisation
Slice:Drop tool is integrated in the cloud-based
image analysis toolbox for image visualization
(Slice:Drop, 2012). It is a viewer for both 2D and
3D biomedical image data, supporting various file
formats. Slice:Drop uses WebGL and HTML5
Canvas to render the image data. Figure 10 shows
the 3D volume rendering of a brain, and its 2D slices
in X, Y and Z directions. The viewer also allows
users to change the intensity threshold to show the
bright areas of interest, and to optimize the display
of the image by adjusting Window/Level of the
volume.
6 CONCLUSIONS AND FUTURE
WORK
In this paper, we have presented the architecture,
design and implementation of the cloud computing
services for biomedical image analysis, which is
running on a national cloud infrastructure provided
as an IaaS. Our aim is to build a cloud enabled
image analysis and processing platform by
integrating a suite of cloud computing components.
The platform provides a development environment
for rapid deployment of image analysis tools. We
have also demonstrated the functionality and usage
of the cloud-based image analysis toolbox and its
applications for biomedical image analysis. Our
preliminary experimental results have shown that the
cloud-based image analysis toolbox offers a
powerful new resource for scientists, due especially
to its scalability, nimbleness and cost-effectiveness.
CLOSER2013-3rdInternationalConferenceonCloudComputingandServicesScience
356

The experiments have shown great promises in
biomedical image analysis applications.
The challenges of the project include the
adaption of existing image processing and analysis
algorithms developed by researchers, visualization
toolkits; and link to online image data repositories.
Our plan for future work includes further improving
the performance of the tools for processing large
scale image datasets, and refining the user interface
with more involvement of the relevant research
communities.
ACKNOWLEDGEMENTS
This work is financially supported by The National
eResearch Collaborative Tools and Resources
(NeCTAR) funding, Australia.
REFERENCES
Monaco, A., 2012. “A View Inside the Cloud”, IEEE
website: http://theinstitute.ieee.org/technology-
focus/technology-topic/a-view-inside-the-cloud.
Pasik, A., 2012. “Cloud Computing: The Inevitable
Answer”, IEEE website, http://theinstitute.ieee.org/
ieee-roundup/opinions/ieee-roundup/cloud-computing-
the-inevitable-answer.
Amazon, 2012. “Amozon Web Services (AWS)”,
http://aws.amazon.com.
Microsoft, 2012. “Windows Azure”, http://
www.microsoft.com/windowazure.
Google, 2012. “Google App Engine”, http://
code.google.com/appengine.
SIMAGIS, 2012. “Cloud Software for Microscopy and
Image Analysis”, http://live.simagis.com/home.
Siemens, 2012. website: http://www.siemens.com/
innovation/apps/pof_microsite/_pof-spring-
2011/_html_en/cloud-computing.html.
PCI Geomatics White Paper, 2011. “On-demand Satellite
Image Processing – Next generation technology for
processing Terabytes of imagery on the Cloud”.
Shams, K., Powell, M., Crockett, T., Norris, J., Rossi, R.,
Soderstrom, T., 2010: “Polyphony: A Workflow
Orchestration Framework for Cloud Computing”,
Proceedings of 10th IEEE/ACM International
Conference on Cluster, Cloud and Grid Computing,
Melbourne, Victoria, Australia, pp. 606-611.
Almeer, M., 2012. “Cloud Hadoop Map Reduce for
Remote Sensing Image Analysis”, Journal of
Emerging Trends in Computing and Information
Sciences, Vol. 3, No. 4, pp. 637-644.
Berriman, G., Deelman, E., Groth, P., and Juve, G., 2010.
“The Application of Cloud Computing to the Creation
of Image Mosaic and Management of Their
Provenance”, SPIE Conference 7740: Software and
Cyberinfrastructure for Astronomy. Editors: N.
Radziwill and A. Bridger.
Golpayegani, N., Halem, M., 2009. “Cloud Computing for
Satellite Data Processing on High End Compute
Clusters”, 2009 IEEE International Conference on
Cloud Computing (CLOUD ’09), Bangalore, India, pp.
88-92.
Soyata, T., Muraleedharan, R., Funai, C., Kwon, M.,
Heinzelman, W., 2012. “Cloud-Vision: Real-time Face
Recognition Using a Mobile-Cloudlet-Cloud
Acceleration Architecture”, Proceedings of 2012 IEEE
Symposium on Computers and Communications
(ISCC), Cappadocia, Turkey, pp. 59-66.
Ferzli R. and Khalife, I., 2011. “Mobile Cloud Computing
Educational Tool for Image/Video Processing
Algorithms”, Proceedings of 2011 IEEE Digital
Signal Procesing Workshop and IEEE Signal
Processing Education Workshop (DSP/SPE), Arizona,
USA, pp. 529-533.
NeCTAR official website, 2012. http://nectar.org.au.
Galaxy website, 2012. http://galaxyproject.org/.
CloudMan website, 2012. http://usecloudman.org/.
Oracle website, 2012. http://www.oracle.com/
us/sun/index.htm.
OpenStack website, 2012. http://www.openstack.org/.
AAF website, 2012. http://www.aaf.edu.au/.
Slice:Drop website, 2012. http://slicedrop.com/.
CloudbasedServicesforBiomedicalImageAnalysis
357
