
Enhancing Collaboration in Big Biomedical Data Settings
Knowledge Visualization, Data Mining and Decision Making Issues
Nikos Karacapilidis
1
, Georgia Tsiliki
2
and Manolis Tzagarakis
1
1
Computer Technology Institute and Press "Diophantus" and University of Patras, 26504 Rio Patras, Greece
2
Biomedical Research Foundation, Academy of Athens, 11527 Athens, Greece
Keywords: Big Data, Data Mining, Decision Support Systems, Collaboration, Knowledge Visualization, Data and
Information Quality, Situational Awareness, Biomedical Data.
Abstract: Biomedical researchers need to efficiently and effectively collaborate and make decisions by meaningfully
assembling, mining and analyzing available large-scale volumes of complex multi-faceted data residing in
different sources. Arguing that dealing with data-intensive and cognitively complex settings is not a
technical problem alone, this paper reports on the development and practical use of an innovative web-based
collaboration support service in a biomedical research context. The proposed service builds on the synergy
between machine and human intelligence to facilitate and augment the underlying knowledge management,
data mining and decision making processes. Evaluation results indicate that the service enables stakeholders
to make more informed decisions, by displaying the aggregated information according to their needs.
1 INTRODUCTION
Collaboration support technologies and platforms
are crucial in today’s biomedical research settings,
where multidisciplinary communities ranging from
biologists to bioinformaticians need to assimilate
clinico-genomic research information and scientific
findings and explore diverse associated issues (Ekins
et al., 2011). At the same time, biomedical research
is associated with large-scale amounts of multiple
types of data, obtained from diverse and distributed
sources. An ever-increasing volume of biomedical
resources, including multiple types of data sets and
analysis tools, are available on the web. For
instance, recent technology advances in Next
Generation Sequencing (NGS) platforms entail an
exponential increase in the size and number of
experimental data sets available (Quail et al., 2012).
However, in most cases, the raw information is so
overwhelming that researchers are often at a loss to
even know where to begin to make sense of it.
This paper reports on a web-based collaboration
support service that aims to fully cover the diversity
of requirements in contemporary biomedical
research settings by providing a series of innovative
features. Firstly, the service provides advanced
collaboration support functionalities through
innovative virtual workspaces based on alternative
data visualizations schemas. Secondly, it is able to
meaningfully accommodate the outcomes of latent
knowledge mining services in a collaboration
session, thus offering added value concerning
recognition of biomedical data patterns. Thirdly, by
supporting emergent semantics and the incremental
formalization of argumentative collaboration, it
augments individual and collective decision making.
By providing ease-of-use and expressiveness for
users and advanced reasoning by the machine, the
service also provides appropriate recommendation
mechanisms that enable stakeholders to project their
future actions in their dynamic working settings.
The proposed platform has been developed in the
context of an FP7 EU research project, namely
Dicode (http://dicode-project.eu/), which exploits
and builds on prominent high-performance
computing paradigms and large data processing
technologies to facilitate and augment collaboration
and decision making in data-intensive and
cognitively-complex settings.
The remainder of this paper is structured as
follows: Section 2 reports on related work and
highlights existing problems and requirements;
Section 3 sketches the overall approach followed in
the Dicode project, putting emphasis on knowledge
visualization, data mining and decision making
issues; Section 4 provides an illustrative example to
demonstrate the use of the proposed service in a
23
Karacapilidis N., Tsiliki G. and Tzagarakis M..
Enhancing Collaboration in Big Biomedical Data Settings - Knowledge Visualization, Data Mining and Decision Making Issues.
DOI: 10.5220/0004492100230031
In Proceedings of the 2nd International Conference on Data Technologies and Applications (DATA-2013), pages 23-31
ISBN: 978-989-8565-67-9
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)

biomedical setting; finally, Section 5 concludes by
discussing related remarks and outlining evaluation
results.
2 RELATED WORK
Easy visualization and analysis of big biomedical
data is a highly important requirement in the settings
under consideration. Towards fulfilling it, a series of
applications and web services that link together
bioinformatic tools and databases have recently
emerged. For instance, BioGRID (Stark et al., 2006),
BNDB (Birkl and Yona, 2006) and BioMart
(Guberman et al., 2011) are repositories which store
readily combined data sets and provide platforms to
easily visualize such data. Similarly, the
GenePattern platform provides access to more than
180 tools for genomic analysis to enable
reproducible in silico research (http://
www.broadinstitute.org/cancer/software/genepattern
/). In addition, many collaborative resource sharing
networks have been established, e.g. the eagle-i
consortium (https://www.eagle-i.net/), to address the
researchers’ data sharing needs and accelerate the
discovery of new knowledge. Integration of these
separate systems and resources into a single flexible
infrastructure that streamlines heterogeneous
workloads is a challenging task.
At the same time, a number of projects and
initiatives aim at addressing diverse collaboration
requirements in a variety of biomedical contexts. For
instance, GRANATUM (http://granatum.org) tries to
bridge the information, knowledge and collaboration
gap by providing integrated access to the globally
available data resources needed to perform complex
cancer chemoprevention experiments and conduct
studies on large-scale datasets; Health-e-Child
(http://www.health-e-child.org) gives clinicians a
comprehensive view of a child’s health by
integrating biomedical data, information and
knowledge that spans the entire spectrum from
imaging to genetic to clinical and epidemiological
data; Virolab (http://www.virolab.org) offers a user
friendly environment to facilitate tasks such as data
archiving, data integration, data mining and
simulation; finally, SIMBioMS (http://
simbioms.org) is a multi-module solution for
biomedical data management that is able to
accommodate experiments requiring non-
conventional data storage solutions.
While certainly helpful in addressing specific
biomedical subjects, the above projects and
initiatives do not deal with big data issues;
moreover, they do not exploit the synergy between
human and machine intelligence in order to
meaningfully accommodate and interpret the results
of the associated data mining services through an
environment that facilitates and enhances
collaboration among stakeholders.
As the number of related Web services is
constantly increasing, their proper integration
becomes a critical issue. A few approaches have
been already launched to facilitate the collaboration,
data sharing and decision making among scientists
by providing them with a platform to share
resources. A well known example of this category of
related work is myExperiment (Goble et al., 2010),
an online research environment that supports the
social sharing of bioinformatics workflows, i.e.
procedures consisting of a series of computational
tasks, which can then be reused according to their
specific requirements. Another representative
example is BioCatalogue (http://
www.biocatalogue.org/), which is a registry of web
services that allows users to annotate and comment
on the available services in order to assist them in
identifying the more suitable ones (services are
presented in terms of their functions, data types and
resources). A third example is MethodBox
(https://www.methodbox.org/), which enables
researchers to browse and download data sets, share
methods and scripts, find fellow researchers with
similar interests and share knowledge. Instead of
workflows, MethodBox users share statistical
methods for epidemiology and public health
research. Finally, the Galaxy Project
(http://galaxy.psu.edu/) offers a web-based platform
allowing researchers to perform and share their
analyses. In any case, approaches of this category
demonstrate a set of limitations, mainly concerning
incorporation of collective intelligence and
flexibility in the integration of services offered.
Moreover, they lack mechanisms for a meaningful
integration of data mining services to appropriately
support tasks such as the discovery of patterns and
dependencies within big data sets, which are very
common in the biomedical research domain.
As results from the above, collaboration in the
biomedical domain involves assembling and
analyzing big volumes of complex multi-faceted
data. In this context, a holistic approach integrating
collaboration, new knowledge co-production,
decision making and data mining services is
required. Biomedical researchers need a highly
flexible service that enables them to easily and
meaningfully embed data mining in their
collaborative data analysis and decision making
DATA2013-2ndInternationalConferenceonDataManagementTechnologiesandApplications
24

process. This service should first of all be focused
on improving efficiency; it has to improve current
work practices that are often characterized by
fragmentation of information and multiple disparate
tools in use; it also has to improve the quality and
speed of the current processes, paying much
attention to data and decision provenance issues.
Secondly, such a service should be focused on
improving effectiveness, in that it enables
stakeholders figuring out how to carry out their daily
tasks better; for instance, how to improve their work
methodologies when elaborating and interpreting big
biomedical data residing in diverse sources. Finally,
such a service should enable stakeholders transform
their work, thus leading to new processes, innovative
work methodologies, and new insights. The solution
described in the next section is geared towards this
direction.
3 COLLABORATION IN DICODE
Dicode provides a novel Web-based collaboration
support service with advanced knowledge
management, data mining and decision making
functionalities. The service enables the seamless
integration of these functionalities and allows their
interoperation from both a technical and conceptual
point of view. In this regard, semantics techniques
have been exploited to define an ontological
framework for capturing and representing the
diverse stakeholder and associated data perspectives.
3.1 Knowledge Visualization Issues
Collaboration in Dicode brings together two
paradigms: the Web 2.0 paradigm, which builds on
flexible rules favouring ease-of-use and human
interpretable semantics, and the traditional decision
support paradigm, which requires rigid rules that
reduce ease-of-use but render machine interpretable
semantics. To achieve this, our approach builds on a
conceptual framework, where formality and the level
of knowledge structuring during collaboration is not
considered as a predefined and rigid property, but
rather as an adaptable aspect that can be modified to
meet the needs of the tasks at hand. By the term
formality, we refer to the rules enforced by the
system, with which all user actions must comply.
Allowing formality to vary within the collaboration
space, incremental formalization, i.e. a stepwise and
controlled evolution from a mere collection of
individual ideas and resources to the production of
highly contextualized and interrelated knowledge
artifacts and finally decisions, can be achieved
(Shipman and McCall, 1994).
Dicode offers alternative visualizations of the
collaboration space (called ‘Dicode views’), which
comply with the incremental formalization concept.
Each Dicode view provides the necessary
mechanisms to support a particular level of
formality. The more informal a view is, the greater
easiness-of-use is implied. At the same time, the
actions that users may perform are intuitive and not
time consuming; however, the overall context is
human (and not system) interpretable. On the other
hand, the more formal a view is, the smaller
easiness-of-use is rendered; the actions permitted are
less and less intuitive and more time consuming. The
overall context in this case is both human and
system interpretable (Karacapilidis and Tzagarakis,
2012). The views that are particularly interesting in
the context of this paper are:
Mind-map View: a collaboration space is
displayed as a mind map (Figure 1), where
users can interact with the items uploaded so
far. The map deploys a spatial metaphor
permitting the easy movement and arrangement
of items on the collaboration space. The aim of
this view is to support information triage
(Marshall and Shipman, 1997), i.e. the process
of sorting and organizing through numerous
relevant materials and organizing them to meet
the task at hand.
Formal View: this view enables the posting of
predefined knowledge items, which adhere to a
specific argumentation model (i.e., IBIS (Kunz
and Rittel, 1970)). It invokes a set of dedicated
scoring and reasoning mechanisms aiming to
aid users conceive the outcome of a particular
collaborative session and receive support
towards reaching a decision (Figure 2).
In the ‘mind-map view’ of the collaboration
space, stakeholders may organize their collaboration
through dedicated item types such as ‘ideas’, ‘notes’,
‘comments’ and ‘services’. Ideas stand for items that
deserve further exploitation; they may correspond to
an alternative solution to the issue under
consideration and they usually trigger the evolution
of the collaboration. Notes are generally considered
as items expressing one’s knowledge about the
overall issue, an already asserted idea or note.
Comments are items that usually express less strong
statements and are uploaded to express some
explanatory text or point to some potentially useful
information. Multimedia resources can also be
uploaded into the mind-map view (the content of
which can be displayed upon request or can be
EnhancingCollaborationinBigBiomedicalDataSettings-KnowledgeVisualization,DataMiningandDecisionMaking
Issues
25

directly embedded in the workspace).
A detailed description of the knowledge
visualization related technologies can be found in
(Karacapilidis et al., 2011).
3.2 Data Mining Issues
In the ‘mind-map view’, service items enable users
to configure, launch and monitor the execution of
external data mining services from within the
collaboration workspace, and allow the automatic
upload of their results into the workspace when the
execution of these services terminates.
As with any other item type, users may specify a
title and a content, which gives the ability to attach a
longer description to the item, when such an item
type is uploaded into the collaboration workspace.
Once uploaded, users may configure the item and
specify which data mining service it corresponds to.
The set of available data mining services with which
the service item can be associated is stored centrally
at a dedicated registry (the exploitation of a specific
data mining service, namely Subgroup Discovery, is
discussed in Section 4). Once service items on the
collaboration workspace have been configured, they
can be executed by supplying the required
parameters to the data mining service. Visual cues
indicate the status of service items: whether they
have yet to be executed, are currently executing or
have finished their execution.
Service items appearing on collaboration
workspaces can be explicitly related with other items
in the workspace via relationships or can be spatially
arranged and grouped. When the execution of a
service terminates, the results – consisting of one or
more files – are automatically uploaded into the
collaboration workspace and explicitly connected to
the service item whose execution produced them.
Once the results of service items are available
(i.e., uploaded into the collaboration workspace),
users can treat them as regular items. Furthermore,
users may rate service items in order to indicate
which service they consider as the most useful one
in the context of the discourse.
More details on the data mining technologies
exploited in our approach can be found in (Tsiliki et
al., 2012).
3.3 Decision Making Issues
In the ‘formal view’ of the collaboration space, the
available knowledge item types include ‘issues’,
‘alternatives’, ‘positions’, and ‘preferences’. Issues
correspond to problems to be solved, decisions to be
made, or goals to be achieved. For each issue, users
may propose alternatives (i.e. solutions to the
problem under consideration) that correspond to
potential choices. Positions are asserted in order to
support the selection of a specific course of action
(alternative), or avert the users’ interest from it by
expressing some objection. A position may also
refer to another (previously asserted) position, thus
arguing in favour or against it.
Finally, preferences provide individuals with a
qualitative way to weigh reasons for and against the
selection of a certain course of action. A preference
is a tuple of the form [position, relation, position],
where the relation can be “more important than” or
“of equal importance to” or “less important than”.
The use of preferences results in the assignment of
various levels of importance to the alternatives in
hand. Like the other discourse elements, they are
subject to further argumentative discourse.
The above four semantic types of items enable
users to contribute their knowledge on the particular
problem or need (by entering issues, alternatives and
positions), as well as to express their relevant values,
interests and expectations (by entering positions and
preferences). Moreover, this view continuously
processes the elements entered by the users (by
triggering its reasoning mechanisms each time a new
element is entered), thus facilitating users to become
aware of the elements for which there is (or there is
not) sufficient (positive or negative) evidence, and
accordingly conduct the discussion in order to reach
consensus.
Alternatives, positions and preferences have an
activation label indicating their current status (they
can be active or inactive). This label is calculated
according to the argumentation underneath and the
type of evidence specified for them. Active elements
are taken into account in a scoring mechanism that
calculates the weight of each alternative expressed
and indicates the one that prevails each time (for
more details on the decision making algorithms
used, see (Karacapilidis and Papadias, 2001)).
Dicode collaboration spaces can be transformed
at any time from one view into another. During such
transformations, the semantically enriched item
types available in one view are transformed into the
respective item types of the desired destination view.
The transformation is rule-based; such rules can be
defined by users participating in a collaboration
session and reflect the evolution of a community’s
collaboration needs. After a transformation into the
desired view occurs, the collaboration may continue
in this view, with the users being able to exploit the
item types available in order to keep conducting the
DATA2013-2ndInternationalConferenceonDataManagementTechnologiesandApplications
26
discourse in the desired formality level and take
advantage of the provided functionality.
4 AN EXAMPLE OF USE
To better illustrate the use of the proposed Web-
based collaboration support service and in particular
how the available functionalities can be used in the
biomedical context, we present a scenario which is
indicative of the way researchers collaborate in this
field, where research is carried out by
multidisciplinary teams consisting of biologists,
medical doctors, clinical researchers and
statisticians, each of which contributes from his/her
perspective to the problem being discussed.
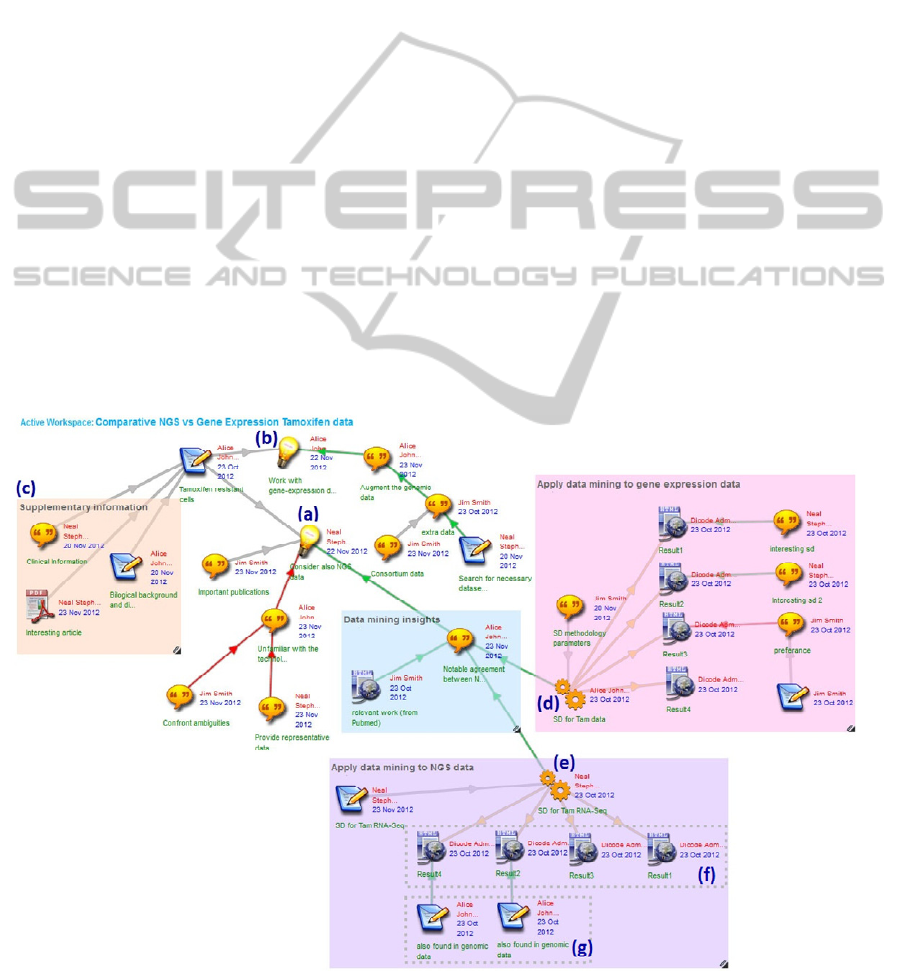
Figure 1 shows the collaboration workspace
operated in the ‘mind-map view’, where a team of
three researchers is discussing an issue related to
breast cancer research. In particular, they are
collaborating in order to determine how to augment
existing datasets in order to study how Tamoxifen
(Tam) resistant cells modulate global gene
expression.
Tam is a widely used antagonist of the estrogen
receptor, whereas its resistance is a well-known
obstacle to successful breast cancer treatment
(Huber-Keener et al., 2012). While adjuvant therapy
with Tam has been shown to significantly decrease
the rate of disease recurrence and mortality,
recurrent disease occurs in one third of patients
treated with Tam within 5 years of therapy. The
team initially selected and analyzed gene-expression
data from 300 patient samples. These data are
derived from whole human genome expression
arrays (Affy U133A Plus 2.0, see
http://www.affymetrix.com). Although the sample is
relatively large, they believe that augmenting the
data with publicly available data will be a good idea
for obtaining statistically significant results.
All participating researchers may upload into the
collaboration workspace items to express their
opinion on the issue being discussed. In the instance
shown in Figure 1, they have uploaded items of type
‘idea’ to propose additional data sets (“Consider also
Next Generation Sequencing (NGS) data” (Figure 1
- (a)) and “Work with gene-expression data (Figure
1 - (b))). Participants have responded to the
proposed alternatives (ideas) by uploading items and
connecting them via arrows to other items to which
they refer. Participants may also change an arrow’s
colour to indicate the semantics of the relationship:
green-coloured arrows express arguments in favour,
red-coloured arrows express arguments against,
Figure 1: Workspace (‘mind-map view’) showing collaboration between biomedical researchers. Service items (d) and (e)
have finished their execution and are associated with other collaboration items.
EnhancingCollaborationinBigBiomedicalDataSettings-KnowledgeVisualization,DataMiningandDecisionMaking
Issues
27
while grey-coloured arrows indicate neutrality.
Furthermore, they can aggregate items on the
workspace by drawing coloured rectangles around
them and give a title to the groupings. For example,
the orange-coloured rectangle with title
“Supplementary information” (Figure 1 - (c)) groups
together bibliographic resources that the team has
obtained from external repositories and are relevant
to their research.
As the discussion evolves, the team thinks about
exploiting the Subgroup Discovery (SD) data mining
algorithm (Atzmueller et al., 2005) using both data
sets as input. SD is the task of finding patterns that
describe subsets of a data set that are highly
correlated relative to a target attribute. This is a
popular approach for identifying interesting patterns
in the data, since it combines a sound statistical
methodology with an understandable representation
of patterns. For example, in a group of patients that
did or did not respond to specific treatment, an
interesting subgroup may be that patients who are
older than 60 years and do not suffer from high
blood pressure respond much better to the treatment
than the average.
To invoke the SD algorithm on the NGS data,
they upload the associated service item into the
workspace (Figure 1 - (e)) and start configuring the
service. Configuring the service includes the
specification of the URI for the REST-based SD
service and specification of parameters such as input
file, number of rules to be used, service ontology,
and minimum number of subgroups to be retrieved
(more details can be found in (Tsiliki et al., 2012)).
After configuring the service, they trigger its
execution. As long as the SD service is executing,
the icon representing the service appears with a
green colour. Upon successful termination of the SD
service, the icon changes its colour to orange and the
results are automatically uploaded into the
collaboration workspace (Figure 1 - (f)). To clearly
indicate the execution of the SD service on the gene
expression data and the results it returned, the team
groups together the relevant items and supplies a
descriptive title (“Apply data mining to NGS data”).
The team can now assess the output of the SD
execution by commenting on the results and
connecting them to other items in the collaboration
workspace (Figure 1 - (g)).
The team can follow the same procedure
(invoking the SD service and collectively assessing
its output) for the gene expression data. The three
researchers are able to carefully examine the
commonalities between the two SD runs (on gene
expression and NGS data) and share their insights.
As the collaboration continues and more items
are added to the collaboration space, the team
decides to switch to a different view, in order to
reach a decision. For this, they decide to transform
the ‘mind-map view’ into the ‘formal view’, which
provides elaborated scoring and reasoning
mechanisms that further facilitate the decision
making process. By transforming the ‘mind-map
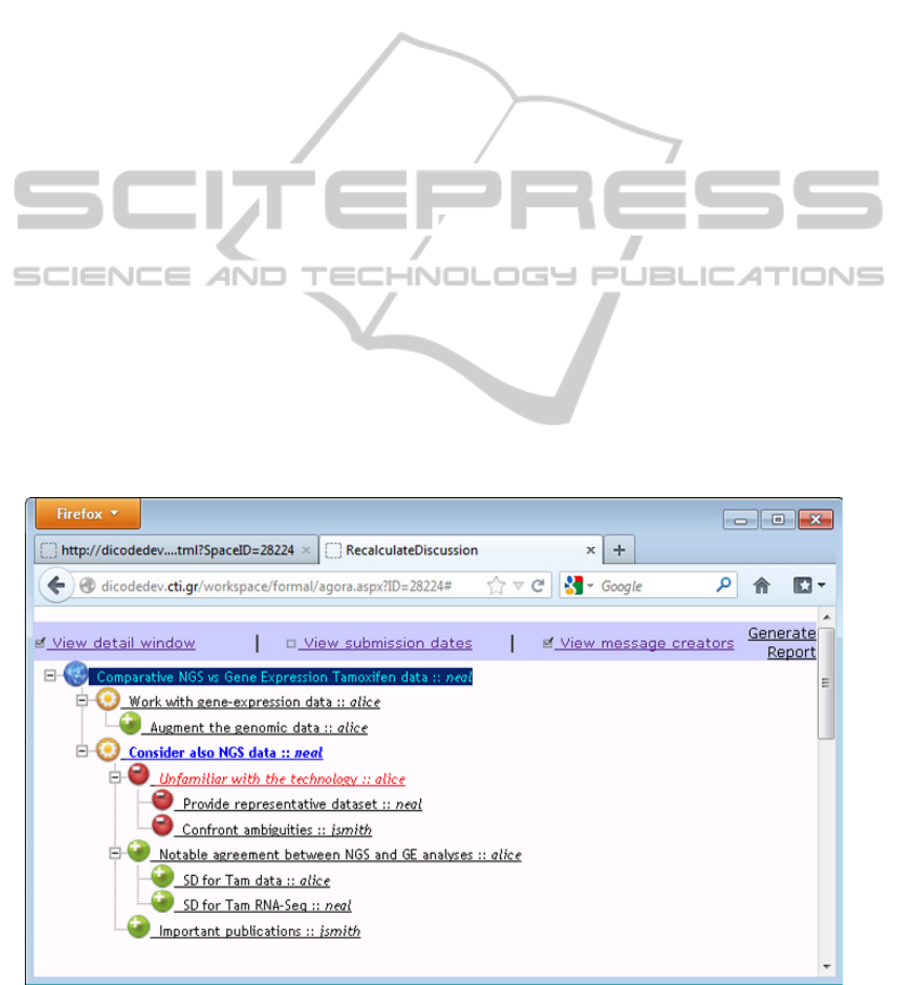
Figure 2: The ‘formal view’ of the collaboration illustrated in Figure 1.
DATA2013-2ndInternationalConferenceonDataManagementTechnologiesandApplications
28
view’ of the collaboration workspace, all available
semantic types – including the service items - are
transformed into the appropriate types of the ‘formal
view’, based on well-specified rules.
Figure 2 shows the collaboration space in the
‘formal view’. As noted above, the team can
continue the collaboration in this view by adding
more items (each time a new item is added, the
reasoning mechanism is triggered). Furthermore, it
allows the team to see which is the best argumented
alternative solution (or ‘winning’ solution) by
highlighting it using visual cues. Based on the
current state of the collaboration, the currently
‘winning’ solution is the alternative “Consider also
NGS data” (item in blue underlined font colour in
Figure 2).
5 DISCUSSION & CONCLUSIONS
5.1 Evaluation Issues
Dicode has been already introduced in three real-life
settings (i.e. the biomedical research assimilator,
decision making on clinical treatment effects, and
opinion mining from unstructured Web 2.0 data) for
a series of pilot experimentations. For the setting
considered in this paper, 61 users from 4 European
countries participated in a detailed evaluation of the
proposed service. The above users had a varying
level of hands-on experience in related technologies
(ranging from ‘early adopters’ to semi-experienced
and novice users); their background was on
disciplines such as Bioinformatics, Biology and
Computer Science. Feedback requested was of both
quantitative and qualitative type. Answers to the
quantitative questions of the questionnaires were
given for ordinal data in a 1-5 scale (questions
concerning the quality, acceptability and
accessibility of the service), where 1 stands for ‘I
strongly disagree’ and 5 for ‘I strongly agree’, and
for continuous numerical data (scale data) in a 0-10
scale (questions concerning the services’ usability),
where 0 stands for ‘none’ and 10 for ‘excellent’.
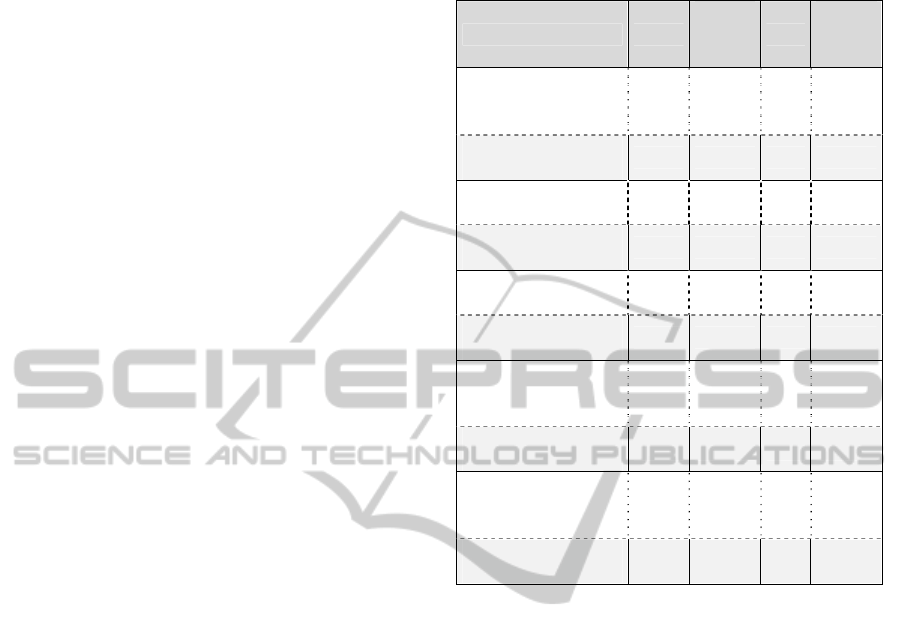
As far as the overall quality of the proposed
collaboration support service is concerned (Table 1),
the evaluators agreed that: the objectives of the
service are met (median=4, mode=3), the service is
novel to their knowledge (median=4, mode=4), they
are satisfied with the performance of the service
(median=4, mode=4), and they are overall satisfied
with this service (median=4, mode=4). The
evaluators seemed to be to some extent sceptical as
Table 1: Overall Quality Descriptive Statistics for the
Dicode Collaboration Support Service.
Question Median
Median
interpret
ation
Mode
Mode
interpret
ation
Q1: The service is able
to address data intensive
decision making issues
3 neutral 3 neutral
Evaluator confidence on
Q1
3 high 2 medium
Q2: The objectives of
the service are met
4 agree 3 neutral
Evaluator confidence on
Q2
3 high 3 high
Q3: The service is novel
to my knowledge
4 agree 4 agree
Evaluator confidence on
Q3
3 high 3 high
Q4: I am satisfied with
the performance of the
service
4 agree 4 agree
Evaluator confidence on
Q4
3 high 3 high
Q5: Overall, I am
satisfied with this
service
4 agree 4 agree
Evaluator confidence on
Q5
3 high 3 high
to whether the service is able to address the data
intensive decision making issues (median=3,
mode=3).
With respect to the acceptability of the service,
the evaluators overall agreed that the service has all
the functionality they expected (median=4,
mode=3), the interface of the service is pleasant
(median=4, mode=4) and that they will recommend
this service to their peers/community (median=4,
mode=3).
The analysis of qualitative evaluation results
showed that, overall, reviewers found the service
“promising”, “easy and intuitive”, as well as “very
useful for a complex use case”. However, a few
technical and documentation issues were raised,
such as: “A bit slow loading time both for the
workspace list and the mind-map view”; “The
arrows’ graphics were not very pleasant for me: they
start from the middle of the icon and not from the
beginning of the square ... the overall idea however,
is quite good”; “I got a bit confused until I fully
understand what I had to do”; “I often missed some
system information”.
Such findings reveal the need for more detailed
documentation of the service, as well as for
provision of help files and system messages.
EnhancingCollaborationinBigBiomedicalDataSettings-KnowledgeVisualization,DataMiningandDecisionMaking
Issues
29

5.2 Final Remarks
The service described in this paper offers an
innovative environment that allows users “immerse”
in Web 2.0 interaction paradigms and exploit its
enormous potential to collaborate through
reviewing, commenting on and extending the shared
content. The Dicode environment enables
stakeholders maintain chains of views and opinions,
accompanied by the supporting data, which may
reflect, at any time, the current collective knowledge
on the issue under consideration, and justify a
particular decision made or action taken.
The proposed service may fully cover the needs
of the three stages of situational awareness needed in
the above settings (Haendel et al., 2012; Kahn,
2011), namely perception (i.e. perceive the status,
attributes, and dynamics of relevant elements in the
setting under consideration), comprehension (i.e.
perform a synthesis of disjointed elements of the
previous stage through the processes of pattern
recognition, interpretation, and evaluation), and
projection (i.e. extrapolate information from
previous stages to find out how it will affect future
instances of the operational setting) (Endsley, 1995).
Moreover, the development of the proposed service
has adopted an agile, analytic and adaptive approach
that enables stakeholders to fully leverage and reap
the benefits of the associated biomedical “big data”.
Such an approach can improve the quality and
effectiveness of decisions in the context under
consideration.
The service described in this paper has been
integrated in the Dicode workbench environment (de
la Calle et al., 2012), which is a web-based
application that integrates - at the level of the user
interface - various data mining and collaboration
support services. The objective is to provide users
with a uniform and easy access to the available
Dicode services. The type and number of services
appearing on the Dicode workbench can be easily
configured by end users according to the needs of
the particular context and problem under
consideration. In such a way, current work practices
have been admittedly improved in terms of
efficiency and effectiveness. The issue of
information fragmentation as well as that of data and
decision provenance are properly addressed.
Moreover, by providing users with useful hints, our
approach enables stakeholders figuring out how to
carry out their daily tasks in a more effective way.
Finally, the proposed service enables stakeholders to
follow and adopt innovative work methodologies,
which build on the synergy of human and machine
reasoning.
Future work directions include investigation of
additional services for data-intensive computing
(e.g. services already developed in projects such as
ADMIRE - http://www.admire-project.eu),
considering whether they can be integrated in the
Dicode environment. Also, a thorough investigation
of the Dataspace concept and the related data
management abstraction (Halevy et al., 2006),
considering its suitability to the purposes of our
approach.
ACKNOWLEDGEMENTS
This publication has been produced in the context of
the EU Collaborative Project “DICODE - Mastering
Data-Intensive Collaboration and Decision”, which
is co-funded by the European Commission under the
contract FP7-ICT-257184. This publication reflects
only the authors’ views and the Community is not
liable for any use that may be made of the
information contained therein.
REFERENCES
Atzmueller, M., Puppe, F. and Buscher, H.P. 2005.
Exploiting background knowledge for knowledge-
intensive subgroup discovery. In Proceedings of
IJCAI’05, 647-652.
Birkl, A., and Yona, G. 2006. Biozon: a hub of
heterogeneous biological data. Nucleic Acids
Research, 34:2006.
de la Calle, G., Alonso-Martinez, E., Tzagarakis, M. and
Karacapilidis, N. 2012. The Dicode Workbench: A
Flexible Framework for the Integration of Information
and Web Services. In Proceedings of the 14th
International Conference on Information Integration
and Web-based Applications & Services (iiWAS2012),
Bali, Indonesia, December 3-5, 2012, pp. 16-25.
Ekins, S., Hupcey, M., Williams, A. (Eds), 2011.
Collaborative Computational Technologies for
Biomedical Research, John Wiley & Sons, Inc.
Endsley, M. R., 1995. Toward a theory of situation
awareness in dynamic systems. Human Factors 37(1),
32–64.
Guberman, J. M., Ai, J., Arnaiz, O., Baran, J., Blake, A.,
Baldock, R., Chelala, C., Croft, D., Cros, A., Cutts, R.
J. et al. 2011. Biomart central portal: an open database
network for the biological community. Database,
2011:bar041.
Goble, C. A, Bhagat, J., Don Cruickshank, S. A.,
Michaelides, D., Newman, D., Borkum, M.,
Bechhofer, S., Roos, M., Li, P. and De Roure, D. D.
2010. myExperiment: a repository and social network
DATA2013-2ndInternationalConferenceonDataManagementTechnologiesandApplications
30

for the sharing of bioinformatics workflows. Nucleic
Acids Research, 38:W677–W682.
Haendel, M. A., Vasilevsky, N. A., Wirz, J. A. 2012.
Dealing with Data: A Case Study on Information and
Data Management Literacy. PLoS Biol. 10, 5,
e1001339.
Halevy, A., Franklin, M. and Maier, D. 2006. Principles of
Dataspace Systems. In Proceedings of the 25th ACM
SIGMOD-SIGACT-SIGART Symposium on Principles
of Database Systems (PODS 2006), Chicago, IL, USA,
pp. 1-9.
Huber-Keener, K. J., Liu, X., Wang, Z. et al. 2012.
Differential Gene Expression in Tamoxifen-Resistant
Breast Cancer Cells Revealed by a New Analytical
Model of RNA-Seq Data. PLoS ONE. 7, 7, e41333.
DOI= doi:10.1371/journal.pone.0041333
Kahn, S. D. 2011. On the future of genomic data. Science.
331, 728, DOI: 10.1126/science.1197891
Karacapilidis, N. and Papadias, D. 2001. Computer
Supported Argumentation and Collaborative Decision
Making: The HERMES system. Information Systems,
26(4), 259-277.
Karacapilidis, N., Karousos, N., Tzagarakis, M. and
Christodoulou, S. 2011. Mitigating the cognitive
overload of contemporary argumentation-based
collaboration settings. In Proceedings of the 7th
International Conference on Collaborative
Computing: Networking, Applications and
Worksharing (CollaborateCom 2011), Orlando, FL,
USA, October 15-18, 2011, pp. 516-519.
Karacapilidis, N. and Tzagarakis, M. 2012. Towards a
Seamless Integration of Human and Machine
Reasoning in Data-Intensive Collaborative Decision
Making Settings: The Dicode Approach. In Proc. of
the 16th IFIP WG8.3 International Conference on
Decision Support Systems (DSS 2012), IOS Press,
Amsterdam, 223-228.
Kunz, W. and Rittel, H. W. J. 1970. Issues as elements of
information systems. Working Paper 131, Institute of
Urban and Regional Development, University of
California.
Marshall, C. and Shipman, F. 1997. Spatial hypertext and
the practice of information triage. In Proc. of the 8th
ACM Conference on Hypertext, Southampton, UK,
124–133.
Quail, A. Q., Smith, M., Coupland, P., Otto T. D., Harris,
S. R., Connor, T.R., Bertoni, A., Swerdlow, H. P., Gu,
Y. 2012. A tale of three generation sequencing
platforms: comparison of Ion Torrent, Pacific
Biosciences and Illumina MiSeq sequencers. BMC
Genom. 13, 431, DOI: 10.1186/1471-2164-13-341.
Shipman, F.M. and McCall, R. 1994. Supporting
knowledge-base evolution with incremental
formalization. In Proc. CHI 94 Conference, 285–291.
Stark, C., Breitkreutz, B. J., Reguly, T., Boucher, L.,
Breitkreutz, A. and Tyers, M. 2006. BioGRID: a
general repository for interaction datasets. Nucleic
Acids Research, 34:D535–D539.
Tsiliki, G., Kossida, S., Friesen, N., Rüping, S.,
Tzagarakis, M. and Karacapilidis, N. 2012. Data
mining based collaborative analysis of microarray
data. In Proceedings of the 24th IEEE International
Conference on Tools with Artificial Intelligence
(ICTAI 2012), Athens, Greece, November 7-9, 2012,
pp. 682-689.
EnhancingCollaborationinBigBiomedicalDataSettings-KnowledgeVisualization,DataMiningandDecisionMaking
Issues
31
