
Patient Identification for Clinical Trials with Ontology-based
Information Extraction from Documents
Peter Geibel
1
, Hebun Erdur
2
, Lothar Zimmermann
1
, Stefan Kr
¨
uger
1
, Kati Jegzentis
3
, Josef Schepers
1
,
Anne Becker
4
, Frank M
¨
uller
1
, Christian Hans Nolte
2
, Jan Friedrich Scheitz
2
, Serdar T
¨
ut
¨
unc
¨
u
2
,
Tatiana Usnich
2
, Markus Frick
4
, Martin Trautwein
4
, Thorsten Schaaf
1
, Alfred Holzgreve
4
,
and Thomas Tolxdorff
1
1
Institute of Medical Informatics, Charit
´
e - Universit
¨
atsmedizin Berlin, Berlin, Germany
2
Department of Neurology (CBF), Charit
´
e - Universit
¨
atsmedizin Berlin, Berlin, Germany
3
Center for Stroke Research Berlin (CSB), Charit
´
e - Universit
¨
atsmedizin Berlin, Berlin, Germany
4
Vivantes - Netzwerk f
¨
ur Gesundheit GmbH, Berlin, Germany
Keywords:
Ontologies, Information Extraction, Computational Linguistics, RDFS, Secondary Use of Health Data,
Patient Recruitment, Clinical Data Warehouse.
Abstract:
In this paper, we describe the use of ontologies in the context of a system for recruiting patients for clinical
trials, which is currently being tested at the Charit
´
e – Universit
¨
atsmedizin Berlin, one of the largest university
hospitals in Europe. The main purpose of the CRDW (Clinical Research Data Warehouse) is to support patient
recruitment for clinical trials based on routine data from the hospital’s clinical information system (CIS). In
contrast to most other systems for similar purposes, the CRDW also makes use of information that is present in
clinical documents like admission reports, radiological findings, and discharge letters. The linguistic analysis
recognizes negated and coordinated phrases. It is supported by clinical domain ontologies that enable the
identification of main terms and their properties, as well as semantic search with synonyms, hypernyms, and
syntactic variants. The focus of this paper is the description of our ontology model, which we tailored to the
particular requirements of our application. In the article, we will also provide an evaluation of the system
based on experimental data obtained from the daily routine work of the study assistants.
1 INTRODUCTION
In this paper, we present research on a software sys-
tem that is currently being developed in a research
project, which is a a collaboration between Charit
´
e
– Universit
¨
atsmedizin Berlin, the largest German uni-
versity hospital, Vivantes – Netzwerk f
¨
ur Gesundheit
GmbH, Germany’s largest state-owned health care
corporation, and an SME software partner. The main
purpose of the CRDW (Clinical Research Data Ware-
house) is to support patient identification for clinical
trials based on routine data from the clinical informa-
tion system (CIS).
In recent years, the secondary use of clinical data
has been considered an important topic of research
since it enables medical progress based on data that
are currently only used for treatment, administra-
tive and billing purposes. Other than our project,
relevant projects in this field include I2B2 (Mur-
phy et al., 2006), EHR4CR (http://www.ehr4cr.eu),
Cloud4Health ( http://www.cloud4health.de/), and
KIS REK (Dugas et al., 2008).
The CRDW allows finding patients that meet the
inclusion and exclusion criteria of clinical trials. The
criteria, for instance, correspond to lab values, pa-
tient data (age, sex), and coded information on di-
agnoses and procedures, i.e., ICD-10 codes for di-
agnoses and OPS codes for procedures. Using also
unstructured data, i.e., doctor’s letters and other clini-
cal documents, allows investigators to formulate more
fine-grained criteria compared to using coded infor-
mation alone. Our system differs from related ap-
proaches by its focus on using computational linguis-
tic methods and on ontology-based information ex-
traction from text documents.
In this paper, we report our experiences in mod-
eling and using ontologies (Staab and Studer, 2009).
These clinical knowledge bases are used by our soft-
230
Geibel P., Erdur H., Zimmermann L., Krüger S., Jegzentis K., Schepers J., Becker A., Müller F., Nolte C., Scheitz J., Tütüncü S., Usnich T., Frick M.,
Trautwein M., Schaaf T., Holzgreve A. and Tolxdorff T..
Patient Identification for Clinical Trials with Ontology-based Information Extraction from Documents.
DOI: 10.5220/0004544702300236
In Proceedings of the International Conference on Knowledge Engineering and Ontology Development (KEOD-2013), pages 230-236
ISBN: 978-989-8565-81-5
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
ware for extracting structured information from texts
(Reeve, 2005; Cowie and Wilks, 2000). We also
present evaluation results that are based on patient
data of the Department of Neurology. For the eval-
uation we considered a series of clinical trials. We
compared the predictions of our system to the assess-
ments of the trial team of the Center for Stroke Re-
search Berlin (CSB), in order to obtain estimates of
precision, recall, and sensitivity.
This paper is structured as follows. After an
overview of the system given in section 2, we de-
scribe the requirements of our specific application and
the chosen ontology model (section 3). Section 4
describes evaluation results of the pilot phase at the
Clinic of Neurology. The conclusions can be found in
section 5.
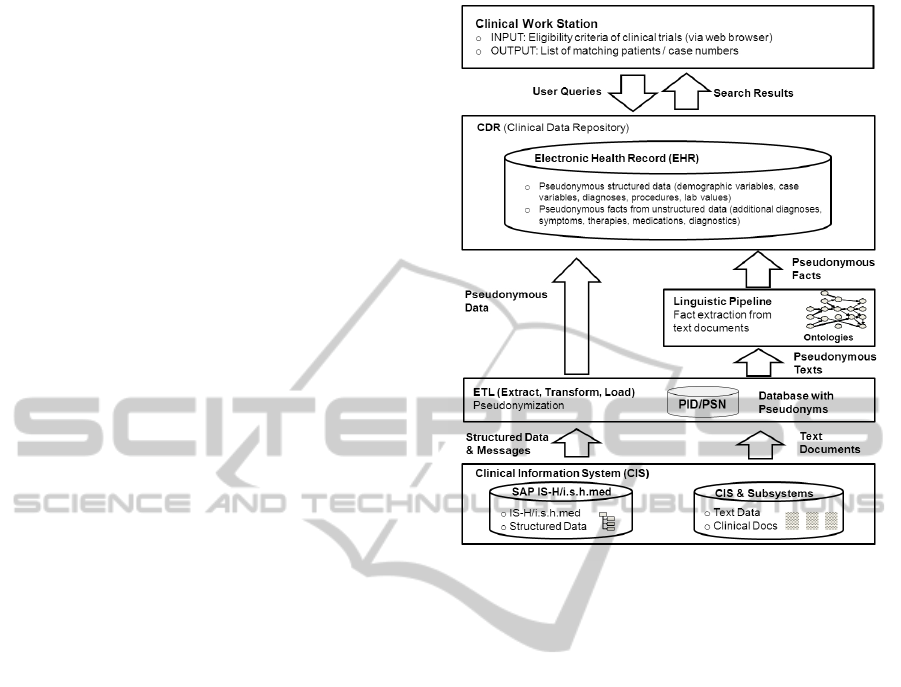
2 OVERVIEW OF THE SYSTEM
In the current version of the CRDW deployed
at Charit
´
e, data from the IS-H/i.s.h.med modules
(SAP/Siemens) of the clinical information system are
integrated with patient information extracted from
documents of the GE radiological system. The struc-
tured data is loaded into the data warehouse by an
ETL (Extract, Transform, Load) process. The data is
stored in a pseudonymous manner in the CDR (clini-
cal data repository), integrating data from both struc-
tured and unstructured sources.
The extraction of patient related facts from text
documents such as radiological findings is based on a
linguistic analysis (M
¨
uller, 2005; Jurafsky and Mar-
tin, 2008), which identifies terms, phrases, and sen-
tences in a text, together with grammatical constructs
such as negation (”not”) and coordination (”and”,
”or”). The identified terms and phrases are then
mapped to medical concepts in a semantic know-
ledge base, which contains etiological, morpholog-
ical, topological and procedural information. This
knowledge allows identifying the main pieces of in-
formation, assigning them to classes like “diagnosis”,
“therapy”, “anatomical structure”, and so on. For in-
stance, in a phrase like “infarction of the MCA”, “in-
farction” will be identified as a diagnosis, whereas
the anatomical structure “MCA” provides informa-
tion about the location of the infarction.
Using the linguistic pipeline together with a know-
ledge base enables semantic search. For instance, the
user can execute queries for synonymous terms like
“stroke” and “cerebral infarction”. The availability
of taxonomical information allows searching for more
general terms with queries like “infarction of a cere-
bral artery”. This query then matches “infarction of
Figure 1: Architecture of the System.
the MCA” and variants hereof like “middle cerebral
artery infarct”. Note that in our approach, ”media in-
farction”, “infarction of the MCA” and “middle cere-
bral artery infarct” are mapped onto the same set of
facts. This cannot be accomplished with a plain text
search.
3 REQUIREMENTS ANALYSIS
AND ONTOLOGY MODEL
In this section, we describe the requirements of our
application, the chosen ontology model, and the
knowledge engineering approach taken.
3.1 Analysis of Clinical Trials
We approached the problem of finding the right on-
tology formalism, structure and content from several
directions. Since the main purpose of the CRDW is
to find patients for clinical trials, we analyzed clini-
cal trials in the field of ischemic strokes, in particu-
lar such that where conducted by the Clinic of Neu-
rology at our hospital. As an example, the clinical
trial TRELAS (Scheitz et al., 2011; Scheitz et al.,
2012) investigates Troponine T elevation in patients
with ischemic stroke. In order to find patients meet-
ing the criteria of this trial, we have to determine pa-
PatientIdentificationforClinicalTrialswithOntology-basedInformationExtractionfromDocuments
231

tients that suffer from a stroke or a non-ST elevation
myocardial infarction (NSTEMI), have a Troponine T
level above 0.05µg/l. Patients with a Creatinine value
above 1.2mg/dl are exluced. Patients with a stroke
can be found by looking at ICD-10 coded diagnoses
in the CIS system, and by analyzing admission reports
and radiological findings, which additionally allows
determining the location of a stroke.
We analyzed stroke studies conducted at Charit
´
e
Berlin with respect to their semantic structure and
typical content. We were able to determine the fol-
lowing groups of criteria relevant for clinical trials for
patients with the diagnosis ”ischemic stroke”:
• Main diagnosis (IS-H/i.s.h.med)
• Age and sex (IS-H/i.s.h.med)
• Radiological report (IS-H/i.s.h.med)
• Symptoms (admission report, discharge letter)
• Lab data (IS-H/i.s.h.med)
• Localization of infarction (radiological reports)
• NIH stroke score (admission report)
• Prior medication, other diseases and therapies
(admission report, previous cases)
• Time of identifying event (admission report)
• Medicolegal aspects as pregnancy, ability to con-
sent, risk factors, are frequently not documented.
As a general result, we were able to verify that
we cannot rely on a single type of information alone
but needed the combination of structured data and
facts extracted from documents. For instance, the
location of a stroke can only be found in text docu-
ments whereas the NIHSS is frequently documented
in the admission report in a structured manner. Both
sources, however, are not 100 % complete, so queries
frequently combine different criteria pertaining to dif-
ferent data sources. Also, part of the information
might not be documented electronically, or might not
yet be available, when searching for eligible patients.
3.2 Mindmaps for Diseases
As a second approach to the question as to how to
design the ontology, we asked the doctors in our
group to draw mindmaps of the diseases, we were
primarily interested in: ischemic stroke, idiopathic
parkinson disease, and multiple sclerosis. For in-
stance, the mindmap of ischemic stroke consists of
the branches: clinical picture, etiology, diagnostics,
differential diagnoses, acute therapy, emergency med-
ical care, complications, outcome, rehabilitation, pre-
vention.
In our requirements analysis, however, we found
that is only necessary to represent the concepts
present in the mindmap, but not its structure given by
the branches and their labels: For instance, we can
can find stroke patients suffering from a paresis by is-
suing the query stroke AND paresis. In order to
be able to answer this query, however, it is not neces-
sary to represent the fact that a paresis is a potential
symptom of a stroke in the ontology.
3.3 Annotation of Clinical Documents
In order to identify the concepts that need to be part of
the ontology, and also in order to be able to construct a
set of test sentences, the clinical doctors were asked to
annotate phrases in a set of example documents cho-
sen by them. In the beginning, they just used a text
marker on a print-out of the documents. Based on the
annotated documents, the ontology models were ex-
tended by a knowledge engineer. In the future, how-
ever, we plan to utilize an annotation tool that allows
the doctors to annotate the relevant phrases graphi-
cally followed by a semi-automatic step of ontology
extension.
3.4 Expressiveness
When starting to develop a prototype of the system,
it was necessary to make a decision for a specific se-
mantic technology. In order to get started, we decided
to set up a SESAME server (http://www.openrdf.org/)
and to use RDFS (RDF Schema) for modeling some
basic concepts.
In RDFS, one can, for instance, establish subclass
relationships between concepts and subproperty rela-
tionships between properties. In our ontologies, we
use the following primitives:
• rdfs:subClassOf for the subclass relationship
• rdfs:subPropertyOf for properties
• rdfs:label for specifying synonyms.
In contrast to synonyms, syntactic variants are fre-
quently handled by pattern matching based on the
phrase structure and the morphological analysis of
words.
Rules. Note that it is not possible in RDFS to de-
fine a concept as the conjunction, disjunction or nega-
tion of other concepts. Neither is it possible to state
rules that derive properties or class membership for
instances.
The lack of rules is partly compensated by the
query interface of the CRDW. In our system, we al-
low conjunction and disjunction of positive criteria
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
232

plus a conjunction of negated criteria. This means
that although we cannot state rules or complex con-
cept definitions, the user can use logical combinations
of search criteria. For instance, instead of inferring
that a patient has diabetes from the fact that he or she
is treated with Metformin, the user of our system can
issue a search for diabetes OR metformin (poten-
tially plus other indicators for diabetes).
As part of our SCRUM (Schwaber and Bee-
dle, 2001) software development process, we care-
fully evaluated several alternatives and extensions
to RDFS: PROLOG (Lloyd, 1987), F-Logic/Object
Logic (Kifer et al., 1995), Datalog (Gallaire et al.,
1984), Production Rules (Drools) (Browne, 2009),
RIF (Kifer, 2008), OWL 2 (Yu, 2011), SPARQL
Rules (SPIN) (Polleres, 2007). Some of these lan-
guages are very powerful but lack built-ins for model-
ing ontologies (e.g., PROLOG, Drools, Datalog). The
remaining approaches pertain to ontologies, however
many of them could not be considered mature enough
to be included into a commercial software product. In
general, we found SPIN the most attractive approach
since it is relatively powerful but lightweight, and it
features negation.
Since there are not may cases, in which rules are
really necessary, though, we postponed the introduc-
tion of SPIN to future versions of our system.
Part-Of. Some of the cerebral arteries have
branches. For instance, the branches of the middle
cerebral artery are called M1, M2, M3, M4. In order
to model this part-of relation and use it for inferenc-
ing, we replaced it with the rdfs:subClassOf rela-
tion, which allows to use the transitive properties of
this relation. Although it is possible in RDFS to state
properties for instances or classes, it is not possible to
define rules for properties (with the exception of using
rdfs:subPropertyOf).
Note that our current approach should be consid-
ered a workaround, which we are planning to replace
by a rule-based approach in the future.
Negation and Uncertainty. The handling of nega-
tion and related issues is a relatively difficult topic
in the clinical context, since diagnoses can be uncer-
tain. The criteria of a clinical trial are divided into
inclusion and exclusion criteria, the latter of which
can be considered negated criteria. Exclusion crite-
ria are usually evaluated in a “closed world manner”.
This means that an exclusion criterion is labeled with
“okay” whenever there is no matching fact that can
be considered certain. In some cases, however, doc-
tors preferred a more conservative “open world ap-
proach”, with exclusion criteria matching only ex-
plicit negative statements. We are therefore planning
on allowing the user to choose between open and
closed world semantics.
A related issue is the handling of uncertain facts
(e.g., suspected diagnoses). For instance, we found it
crucial that the system avoids false negatives, i.e. pa-
tients that are not suggested for a trial although they
are potential candidates. This means that if an exclu-
sion criterion matches an uncertain fact, the respec-
tive patient should still be suggested to the user, i.e.,
the exclusion criterion should not match. In contrast,
inclusion criteria are (usually) required to match also
unsure facts. Since this is not always the case, though,
we are planning on letting the user decide the match-
ing behaviour of each criterion.
3.5 Ontology Structure
The structure of the ontology is basically determined
by the recognition process, which first tries to identify
so-called main terms corresponding to diagnoses and
therapies, and in a second step attaches properties to
these main terms. The main classes of the ontology
are
• Observation: Main term class for diagnoses,
symptoms, and clinical findings.
• Therapy: Main term class for therapeutic proce-
dures including medications
• Anatomy: Class for anatomical entities. Part-of is
represented by rdfs:subClassOf.
• Attributes: Attributes for observations
and therapies. Examples are left, right,
parietal, frontal, acute, chronic. Other
attributes are diagnostic procedures that might be
attached as properties to diagnoses or therapies
3.6 Meta-modeling
In order to help the concept mapping algorithm,
which attaches properties to main terms, we decided
to specify possible attributes for each concept in order
to reduce ambiguities. Consider, for instance, the
sentence ”acute MCA infarction”. In the ontology,
infarction is specified to have potential attributes
”:Acute” and :ArteriaCerebriMedia. This is
achieved by the following declaration:
:Infarction rdf:type rdfs:Class ;
rdfs:subClassOf :Observation;
:label
preferred "Infarction" ;
:hasLocalisation :Artery ,
:Brain ,
:Heart ;
PatientIdentificationforClinicalTrialswithOntology-basedInformationExtractionfromDocuments
233

:hasAttribute :Position ,
:InfarctionAttributes .
The possibility to attach :Acute is inherited from
a superclass of :Infarction. Note that, although
meta-modeling is allowed in RDFS, inheritance of
class relations is not part of the RDFS specification.
Inheriting meta-relations is thus an addition we made
for the project.
3.7 Collaboration and Modularization
At the moment, the ontologies are being developed
by several people who work at different locations and
have different backgrounds. Yet, there is a strong
overlap between ontologies for separate diseases be-
cause co-morbidity has also to be modeled to some
extend, and anatomical and attributive information
have to be shared between ontologies. For now, we
decided to use independent ontology modules. In
order to be able to use different modules in paral-
lel, we plan to use techniques of ontology alignment
(Shvaiko and Euzenat, 2011; Todorov et al., 2010).
3.8 Technical Issues
Based on the requirements analysis described in the
last section, we modeled the ontologies in our project
using RDFS (RDF Schema). We use a triple store,
SESAME, for storing the ontology. Since we found
triple store based inference too slow for or purposes,
the data, however, are stored in a NoSQL database.
When querying the database, the technique of query
expansion is used in order to allow automatic infer-
ences with respect to class and property hierarchy.
3.9 Available Clinical Ontologies
In our project, we also investigated if we can use al-
ready existing knowledge bases.
UMLS (Unified Medical Language System) (Boden-
reider, 2004) is a so-called meta-thesaurus, which
combines several thesauri by the means of a common
semantic network. Since most of the resources are not
available in German, and license conditions are fre-
quently problematic for the use in a commercial soft-
ware, we were not able to use this powerful resource
in our project. UMLS is also used for the linguistic
component in I2B2 (Murphy et al., 2006), a system
that has a similar purpose as the CRDW.
MESH (Medical Subject Headings) (Rogers, 1963)
is a controlled vocabulary for indexing the MED-
LINE/PUBMED database. There exists also a Ger-
man version (MESH GER), which we licensed for the
project. However, in the end we did no use it in our
software since the concepts do not meet the require-
ments of an ontology and the overall structure was not
consistent with our modeling strategy.
As an example, there is a concept called ”In-
farkt, A. cerebri media” in MESH GER. This concept
comprises non-synonymous labels like ”A.-cerebri-
media-Syndrom”, ”A.-cerebri-media-Embolus”, ”A.-
cerebri-media-Thrombose”, ”Left Middle Cerebral
Media Infarction”, ”Right Middle Cerebral Media In-
farction” and variants of ”Infarkt, Arteria cerebri me-
dia”. We also found that many concepts relevant for
us are missing in MESH. In addition, we wanted to
treat a diagnosis and its location as separate concepts.
Because of these reasons, we favored modeling the
ontology from scratch with the help of domain ex-
perts.
OpenGalen: The GALEN Common Reference
Model (CRM) is a clinical terminology, which was
developed in a project funded by the European Union.
The English version is available as an OWL down-
load whereas we could not find any German version.
In general, we found the structure of the GALEN
common reference model much too complex for our
project. We had the feeling that constructing a sim-
pler ontology from scratch is preferable to adapting
the structure of the GALEN model for our purposes.
SNOMED/SNOMED CT: SNOMED CT (System-
atized Nomenclature of Medicine – Clinical Terms,
(Ruch et al., 2008)) is a well-known health care ter-
minology. SNOMED CT consists of a “IS A” hier-
archy along with the possibility to define concepts
based on attributes. SNOMED, the predecessor of
SNOMED CT, was defined using 11 groups of con-
cepts. Since there is no (available) German version of
either SNOMED or SNOMED CT, we were not able
not use it in our project. The same reason prevented
us from using the Foundational Model of Anatomy
(FMA, (Rosse and Mejino, 2003)) and RADLEX.
ICD-10: The “International Statistical Classification
of Diseases and Related Health Problems, 10th Re-
vision” (ICD-10) is the most important classification
of diagnoses. It is widely used for billing purposes
in German hospitals. Although the ICD-10 is quite
broad on the one hand, it is not fine-grained enough
for our purposes: for instance, with respect to the di-
agnosis “stroke”, one is usually interested in the spe-
cific location of the stroke. However, the ICD-10 only
distinguishes between cerebral and pre-cerebral arter-
ies. OPS is used for coding therapies and thus plays
a similar role as ICD-10.
Both ICD-10 and OPS are available in our soft-
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
234
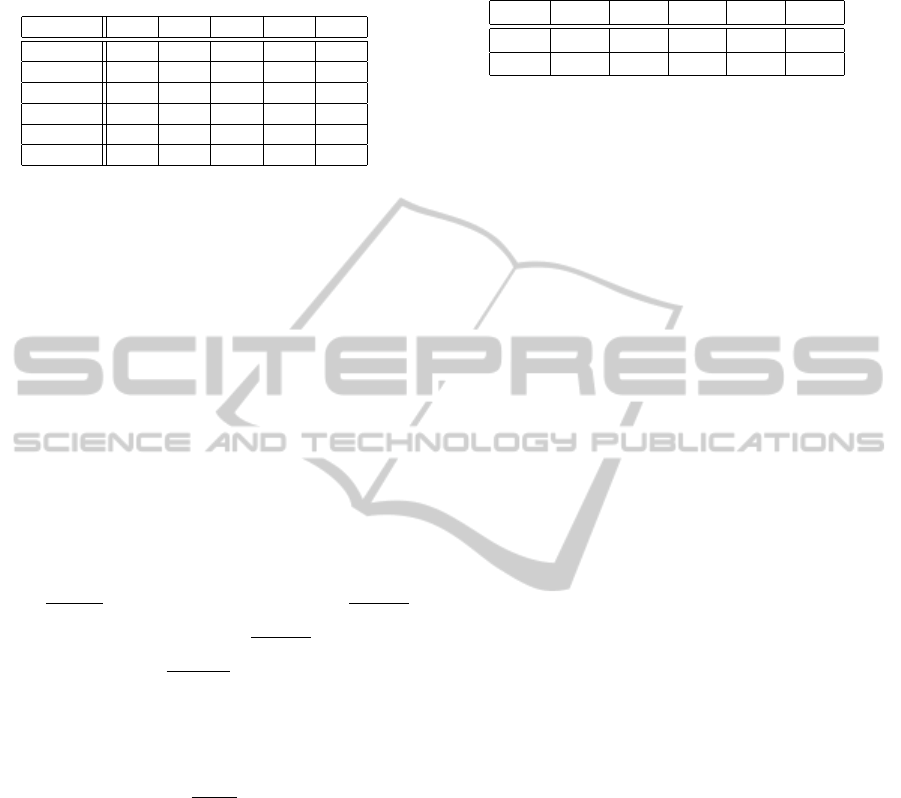
Table 1: System performance for several clinical trials (pre-
cision (positive predictive value), recall (sensitivity), speci-
ficity, negative predictive value, F-measures F).
Trial P R S N F
Trial 2 0.24 0.80 0.39 0.89 0.36
Trial 6 0.36 0.82 0.53 0.90 0.50
Trial 7 0.27 0.80 0.40 0.88 0.40
Trial 8 0.44 1.00 0.91 1.00 0.61
Trial 9 0.33 1.00 0.89 1.00 0.49
Trial 14 0.31 0.91 0.40 0.94 0.46
ware for searching structured data. However, we do
not use it for extracting information from texts.
4 EXPERIMENTS
In the evaluation, we considered 16 stroke Trials,
which are currently being conducted at the Clinic
of Neurology, and compared the performance of the
CRDW to that of the trial team, whose assessment of
patients was considered the gold standard. The data
were collected from January to March 2013. For the
evaluation, we considered only trials with more than
10 candidates in order to obtain more reliable results.
Table 1 shows the evaluation results when using
both structured and unstructured data. For the 6 re-
maining neurological trials, the table shows the pre-
cision
TP
TP+FP
(positive predictive value),
TP
TP+FN
recall (sensitivity), specificity
TN
TN+FP
, the nega-
tive predictive value
TN
TN+FN
plus the so-called f-
measure, (with true positives TP, false positives FP,
true negatives TN, false negatives FN). The F-
measure is the harmonic mean of precision and recall.
It is defined as
F =
P · R
P + R
.
The table shows that we could really achieve a
good sensitivity for all studies, which was of utter-
most importance for the task of patient recruitment.
This means that the system suggests most of the eli-
gible patients. Only among the suggested patients the
study assistant has to determine those patients, which
are actually meeting the criteria of the trial. This en-
hances the time efficiency of the screening process.
Compared to recall, the precision attains lower
values, meaning that the system tends to incorrectly
suggest patients as candidates, reducing the potential
amount of time that can be saved when working with
the system. One problem regarding precision is that
not all necessary information is documented in the
clinical information system. Some information is just
missing or incomplete (e.g., NIH stroke score, med-
Table 2: Precision, recall, specificity, negative predictive
value, F (averages over all trials).
P R S N F
S+U 0.32 0.89 0.59 0.94 0.47
S 0.31 0.81 0.54 0.92 0.43
ications) due to the work load in the ER. Other in-
formation can only be obtained by talking to the pa-
tient or by further examinations. This means that it is
not possible to attain a precision of 1.0. In order to
still improve the performance of the CRDW, we are
currently in the process of increasing the logical ex-
pressiveness of our query interface, which does not
correspond to full SPARQL yet.
In order to determine the usefulness of our
ontology-based approach, we considered two variants
of the data and the study criteria:
• S+U: This corresponds to the complete data set,
comprising unstructured data (U) as well as struc-
tured data (S). For instance, Trial 13 has a dis-
junctive criterion SensorySymptom OR NIHSS-8
>= 1. This means that a sensory symptom has
to be present in some document (e.g., the text
fields of the admission report) whereas the par-
tial stroke score NIHSS-8 is available as a struc-
tured data item. Since NIHSS-8 also pertains to
sensory symptoms, the disjunction expresses in a
redundant manner that a sensory symptom has to
be present. The redundancy is helpful since both
the documentation of the stroke score and the text
sources are not 100% reliable.
• U: Unstructured data only, e.g. NIHSS-8 >= 1
The trial criteria for “S+U” were defined by med-
ical doctors, who are domain experts. The criteria for
’S’ were obtained by removing conditions that pertain
to information contained in documents.
The table fig. 2 shows precision, recall, speci-
ficity, negative predictive value, and F-measure av-
eraged over all 6 trials. Using both structured and
unstructured data results in a high recall and a good
specificity. Dropping conditions from the criteria that
pertain to texts on average results in lower recall, pre-
cision, specificity, negative predictive value, and F-
Measure.
Since the number of trials, 6, is relatively small,
we could not show that the differences are statistically
significant. However there seems to be a trend that the
usage of information extracted from texts increases
the recall, and might also increase specificity.
PatientIdentificationforClinicalTrialswithOntology-basedInformationExtractionfromDocuments
235

5 CONCLUSIONS
In this paper, we described a case study in using on-
tologies for information extraction from clinical doc-
uments. We demonstrated that we managed to build
a system with a high sensitivity – a requirement for
the task of patient recruitment. Improving precision,
however, is still an issue. Future work will focus on
the elimination of false positives by allowing to con-
struct logically more complex criteria.
The experimental data suggest that the process
of patient identification benefits from extracting facts
from structured data. We are planning to obtain more
reliable results by considering more patients and tri-
als. Moreover, the software will be tested by other
departments, too.
A lesson learned in the area of ontologies is that
it can be much easier to construct an ontology for a
specific application instead of building or even using
a general-purpose ontology. However, we also feel
that the lack of German language resources hinders
progress in the domain of semantic technologies suit-
able for German text and web resources.
ACKNOWLEDGEMENTS
The BFG project is partially funded by TSB Tech-
nologiestiftung Berlin, Zukunftsfonds Berlin, and co-
financed by the European Union – European Fund for
Regional Development.
We would like to thank all computer scientists, lin-
guists, ontologists, medical doctors, study nurses, and
administrative staff who participated in the develop-
ment of the software.
REFERENCES
Bodenreider, O. (2004). The unified medical language sys-
tem (umls): integrating biomedical terminology. Nu-
cleic Acids Research, 32(Database-Issue):267–270.
Browne, P. (2009). Jboss Drools Business Rules. From
technologies to solutions. Packt Publishing, Limited.
Cowie, J. and Wilks, Y. (2000). Information extraction.
Handbook of Natural Lang. Proc., pages 241–260.
Dugas, M., Lange, M., Berdel, W., and M
¨
uller-Tidow, C.
(2008). Workflow to improve patient recruitment for
clinical trials within hospital information systems - a
case-study. Trials, 9(1):2.
Gallaire, H., Minker, J., and Nicolas, J.-M. (1984). Logic
and databases: A deductive approach. ACM Comput.
Surv., 16(2):153–185.
Jurafsky, D. and Martin, J. H. (2008). Speech and Language
Processing (2nd Edition) (Prentice Hall Series in Ar-
tificial Intelligence). Prentice Hall, 2 edition.
Kifer, M. (2008). Rule interchange format: The framework.
In Web Reasoning and Rule Systems, volume 5341 of
LLNCS, pages 1 – 11.
Kifer, M., Lausen, G., and Wu, J. (1995). Logical foun-
dations of object-oriented and frame-based languages.
Journal of the ACM, 42(4):741–843.
Lloyd, J. W. (1987). Foundations of Logic Programming,
2nd Edition. Springer.
M
¨
uller, F. (2005). A finite-state approach to shallow pars-
ing and grammatical functions annotation of German.
PhD thesis, University of T
¨
ubingen.
Murphy, S. N., Mendis, M. E., Berkowitz, D. A., and
Chueh, I. K. H. (2006). Integration of clinical and
genetic data in the i2b2 architecture. In AMIA Annu
Symp Proc, page 2009.
Polleres, A. (2007). From SPARQL to rules (and back).
In Williamson, C. L., Zurko, M. E., Patel-Schneider,
P. F., and Shenoy, P. J., editors, WWW, pages 787–796.
ACM.
Reeve, L. (2005). Survey of semantic annotation platforms.
In Proceedings of the 2005 ACM Symposium on Ap-
plied Computing, pages 1634–1638. ACM Press.
Rogers, F. B. (1963). Medical subject headings. Bull Med
Libr Assoc, 51:114 – 116.
Rosse, C. and Mejino, J. (2003). A reference ontology
for biomedical informatics: the foundational model of
anatomy. J Biomed Inform, 36:478–500.
Ruch, P., Gobeill, J., Lovis, C., and Geissb
¨
uhler, A. (2008).
Automatic medical encoding with SNOMED cate-
gories. BMC Medical Inf. and Dec. Making, 8:6.
Scheitz, J. F., Mochmann, H. C., Fiebach, B. W. B., Aude-
bert, H. J., and Nolte, C. H. (2012). J Neurol, 25.
Scheitz, J. F., Mochmann, H. C., Nolte, C. H., Haeusler,
K. G., Audebert, H. J., Heuschmann, P. U., Laufs, U.,
Witzenbichler, B., Schultheiss, H. P., and Endres, M.
(2011). Troponin elevation in acute ischemic stroke
(TRELAS) – protocol of a prospective observational
trial. M. BMC Neurol, 11(98).
Schwaber, K. and Beedle, M. (2001). Agile Software Devel-
opment with Scrum. Prentice Hall PTR, Upper Saddle
River, NJ, USA, 1st edition.
Shvaiko, P. and Euzenat, J. (2011). Ontology matching:
State of the art and future chall. IEEE TKDE, 99.
Staab, S. and Studer, R. (2009). Handbook on Ontologies.
Springer, 2nd edition.
Todorov, K., Geibel, P., and K
¨
uhnberger, K.-U. (2010).
Mining concept similarities for heterogeneous ontolo-
gies. In Perner, P., editor, Advances in Data Mining.
Applications and Theoretical Aspects, volume 6171 of
LNCS, pages 86–100. Springer Berlin / Heidelberg.
Yu, L. (2011). A Developers Guide the Semantic Web.
Springer.
KEOD2013-InternationalConferenceonKnowledgeEngineeringandOntologyDevelopment
236
