
Semantic-based Knowledge Discovery in Biomedical Literature
Fatiha Boubekeur
1
, Sabrina Cherdioui
1
and Yassine Djouadi
2
1
Department of Computer Science, Mouloud Mammeri University, Tizi-Ouzou, Algeria
2
Department of Computer Science, USTHB University, Algiers, Algeria
Keywords: Knowledge Discovery, Biomedical Literature, MEDLINE, MeSH Concepts, Semantic Relatedness,
Swanson’s Discovery.
Abstract: Knowledge discovery in literature aims at searching for hidden and previously unknown knowledge within
the published literature. Swanson’s discoveries on fish oil/Raynaud disease or migraine/magnesium
connections from MEDLINE, an online bibliographic biomedical database, exemplify such discovery. In
this paper, we present a novel approach for literature-based knowledge discovery that relies on the joint use
of (1) flexible information retrieval techniques and (2) concepts’ semantic relatedness to discover hidden
connections between MeSH concepts in the published biomedical scientific literature. The approach has
been tested by replicating the Swanson's early discovery on fish oil/Raynaud disease connection. The
obtained results show the effectiveness of our approach.
1 INTRODUCTION
Literature-Based Discovery (LBD) has been
proposed by Don Swanson (Swanson et al., 1986a)
as a means to identify latent connections between
concepts of interest, that have not been previously
explicitly stated in the published literature (Ganiz et
al., 2005). For this aim, in most LBD approaches,
the literature associated with each concept of interest
is first extracted from an online database, and then
indexed with concepts. The existence of shared
concepts between two literatures reveals a
correlation between the corresponding concepts of
interest.
A key characteristic of these approaches is that
the correlation between two concepts depends on the
existence of shared concepts between their
respective literatures. Therefore, in such “lexical
focused” approaches, correlations are not discovered
between two given concepts if the corresponding
literatures do not share concepts in common, even if
they contain semantically related ones.
To address this shortcoming, in this paper we
propose a novel “semantic focused” approach for
discovering hidden connections between MeSH
concepts in the published biomedical scientific
literature, that relies on concepts semantic
relatedness. In our proposed approach, concepts of
interest are first used to retrieve the corresponding
literatures from MEDLINE database, documents of
which are then indexed using MeSH concepts. The
intersection of these literatures is then examined
through the existence or not of semantically related
concepts. This "semantic intersection" aims at
finding hidden correlations between concepts of
interest that a classical "lexical intersection" does
not succeed to find.
The remainder of the paper is structured as
follows: Section 2 introduces background
knowledge about LBD and biomedical resources,
namely MEDLINE/PubMed and MeSH thesaurus.
Section 3 presents some representative works in
LBD and situates our contribution. Section 4 details
our semantic-based LBD approach. Experimental
results are presented in section 5. Section 6
concludes the paper.
2 BACKGROUND
2.1 What is LBD?
LBD was first proposed by Don R. Swanson in his
pioneering works (Swanson, 1986a; 1986b) as a
solution to discover implicit connections among
information contained in the published biomedical
literature. In his so-called open discovery model
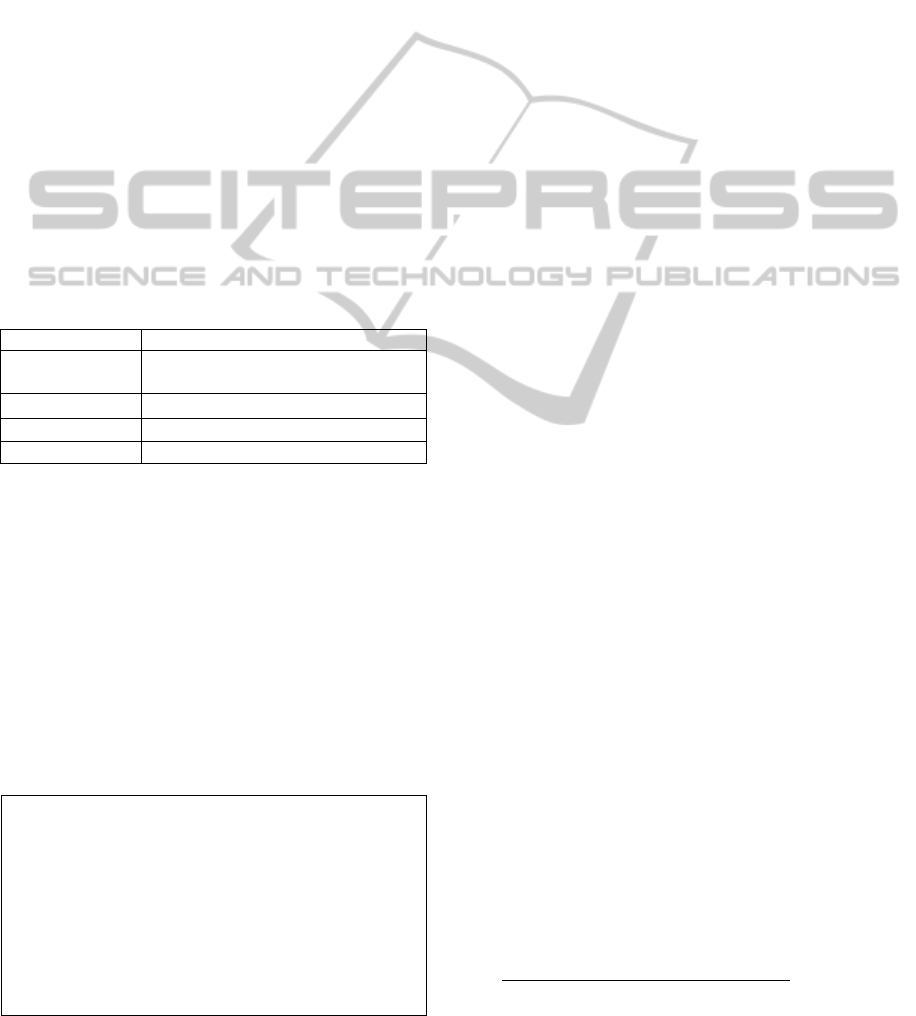
(Figure 1), also known as ABC or ABC model,
the discovery process begins with a starting concept
37
Boubekeur F., Cherdioui S. and Djouadi Y..
Semantic-based Knowledge Discovery in Biomedical Literature.
DOI: 10.5220/0004546300370044
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval and the International Conference on Knowledge
Management and Information Sharing (KDIR-2013), pages 37-44
ISBN: 978-989-8565-75-4
Copyright
c
2013 SCITEPRESS (Science and Technology Publications, Lda.)
A, also called A-concept, related to a research
question. This concept is used to query MEDLINE
online database. The set of all documents that
contain A-concept are retrieved in response to this
query. This set of retrieved documents is called start-
literature or A-literature. Important concepts are then
manually extracted from A-literature. These
concepts are called intermediate concepts or B-
concepts. B-concepts are again used to query the
online database; the resulting document set is the
intermediate literature or B-literature. The
intermediate literature is then processed and
important concepts, deemed the target concepts or
C-Concepts, are selected (e.g. if A refers to an
activate substance such as drug, chemical substance,
proteins and so on, then B might be a physiological
function, a symptom, …and C a pathology or
disease). If concepts A and C do not appear together
in the literature, one has discovered a new (latent)
connection between A and C through the common
B-concepts. This revealed connection is a potential
hypothesis that should be confirmed or rejected by
human experts.
Figure 1: Open discovery (Liu and Fu, 2012).
Based on his ABC model, Swanson has
contributed several major discoveries, from which
his first discovery (Swanson, 1986a; 1986b) on the
connection between fish oil (A-concept) and
Raynaud's disease (C-concept) through the common
physiological B-concepts blood viscosity, platelet
aggregation and vascular reactivity; or the
connection between of magnesium deficiency and
migraine (Swanson, 1988;1989).
Relying on a different discovery approach,
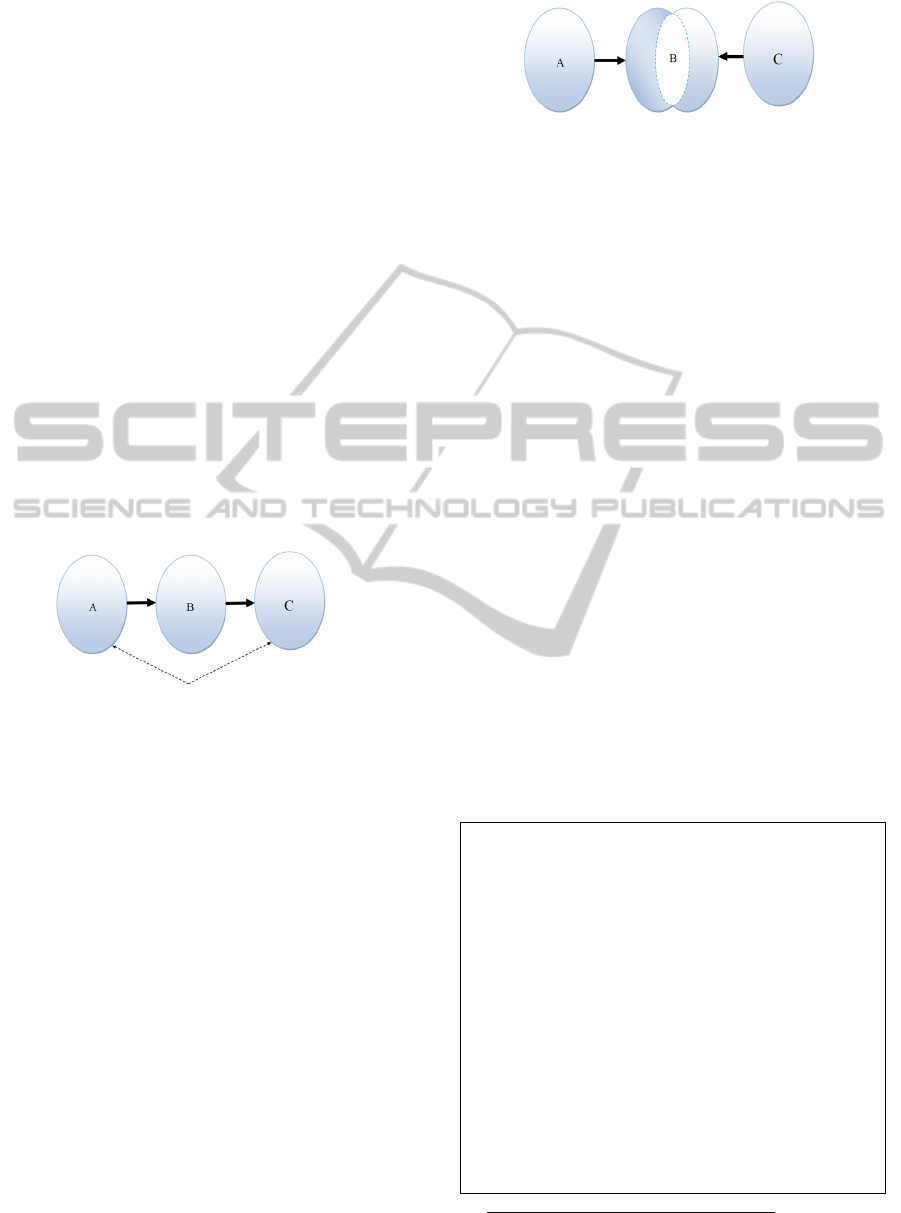
closed discovery models (Figure 2), or ABC
models (Weeber et al., 2001) aim at explaining an
initial observation or hypothesis on a potential
correlation between two given concepts A and C.
Starting with A- and C-literatures, the models
explore the potential connections between A and C
concepts through the existence of common B-
concepts between the corresponding A- and C-
literature’s.
Figure 2: Closed discovery (Liu et al., 2012).
2.2 Biomedical Resources
In this section, we briefly introduce the main
biomedical resources used in our approach.
2.2.1 MEDLINE/PubMed
MEDLINE is the most important biomedical
bibliographic database. It is freely accessible online
via the Entrez information retrieval system,
PubMed
1
.
MEDLINE contains more than 22 million
records of articles from leading biomedical journals.
Each MEDLINE record contains citation, abstract (if
available) and MeSH (Medical Subject Headings)
(Coletti et al., 2001) indexing terms corresponding
to a particular article. An example of MEDLINE
record displayed by PubMed is presented in Table 1,
where TI, AB and MH respectively stand for title,
abstract and MeSH terms corresponding to the
article.
PubMed/MEDLINE’s information retrieval (IR)
model is based on a strict boolean search without
ranking. Therefore, PubMed returns a huge amount
of biomedical documents without being able to rank
them according to their relevance.
Table 1: A Pubmed displayed MEDLINE record.
TI - The effects of dietary omega-3 polyunsaturated fatty
acids on erythrocyte membrane phospholipids, erythrocyte
deformability and blood viscosity in healthy volunteers.
AB - We have examined, in normal subjects, the effects o
f
a daily dietary supplement of fish oil concentrate ('max
EPA'), providing 3 g of omega-3 fatty acids, o
n
erythrocyte membrane phosphor-lipids, erythrocyte
deformability and blood viscosity.
MH - Blood Viscosity/*drug effects
MH - *Docosahexaenoic Acids
MH - Drug Combinations
MH - *Eicosapentaenoic Acid
MH - Erythrocyte Deformability/*drug effects
MH - Erythrocyte Membrane/analysis/*drug effects
MH - Fatty Acids/blood
MH -
…
1
http://www.pubmed.gov
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
38
2.2.2 MeSH Thesaurus
MeSH (Coletti et al., 2001) is the U.S. NLM’s
(National Library of Medicine) controlled
vocabulary thesaurus used for indexing biomedical
articles for the MEDLINE database. It consists of
about 26,853 main headings (or descriptors)
representing concepts found in the biomedical
literature.
A MeSH concept consists of one or several
synonymous terms, one of which is the preferred
term.
A MeSH heading (or MeSH descriptor)
is
formed of one or several related concepts, one of
which is the preferred concept. Other subordinates
concepts are semantically related to the preferred
concept, either through hierarchical relationships,
traditionally thought of as broader/narrower
relationships (such as is-a or part-of), or through
associative (non-hierarchical) relationships (such as
is-caused-by). An example of MeSH heading is
depicted in Table 2.
Table 2: MeSH heading example.
MeSH Heading Blood Platelets
Tree Number A11.118.188
A15.145.229.188
Preferred Concept Blood Platelets
Entry Term
Platelets
Entry Term Thrombocytes
MeSH descriptors are organized into 16 main
fields (or categories) ranging from Anatomy [A],
Organisms [B], Diseases [C], Chemicals and Drugs
[D], Analytical, Diagnostic and Therapeutic
Techniques and Equipment [E], … Each category is
structured into a hierarchy. A given descriptor may
appear at several locations in the hierarchical tree.
The tree locations carry systematic labels known as
tree numbers, and consequently one descriptor can
carry several tree numbers. Table 3 shows an excerpt
from the hierarchy of MeSH’s Diseases category.
Table 3: A MeSH sub-hierarchy.
Diseases [C]
…
Cardiovascular Diseases [C14]
Vascular Diseases [C14.907]
Peripheral Vascular Diseases [C14.907.617]
Blue Toe Syndrome [C14.907.617.249]
Erythromelalgia [C14.907.617.500]
Livedo Reticularis [C14.907.617.625]
Peripheral Arterial Disease [C14.907.617.671]
Phlebitis [C14.907.617.718] + …
3 RELATED WORK
Most of LBD works attempt to replicate Swanson’s
early discoveries on fish oil/Raynaud disease
(Swanson, 1986b), or on migraine/magnesium
deficiency connections (Swanson, 1988), through
either open or closed discovery approaches.
3.1 Open LBD Approaches
Existing open LBD approaches, inspired from
Swanson’s ABC model, rely on various automatic
text indexing techniques to represent the literatures
of interest with accurate features. In (Gordon et al.,
1996; 1999), A-literature is indexed with mono- and
bi-gram keywords that are weighted using a classical
TF*IDF weighting scheme. More advanced
approaches (Gordon and Dumais, 1998; Weeber et
al., 2001; Srinivasan, 2004; Hu et al., 2010; Chen et
al., 2011) base on concepts to index the A-literature.
The indexing concepts are identified either from the
document content using a latent semantic indexing
(LSI) technique (Gordon and Dumais, 1998), or
from a biomedical resource such as MeSH thesaurus
(Chen et al, 2011; Hu et al., 2010; Srinivasan, 2004)
or UMLS
2
(Unified Medical Language Systems)
meta-thesaurus (Weeber et al., 2001), using a
document-ontology mapping technique. Moreover,
in (Srinivasan, 2004), MeSH terms (representing
MeSH concepts) are grouped into vectors according
to their UMLS semantic types (-UMLS organizes
MeSH terms into 134 semantic types or categories-).
The set of all vectors thus obtained is the MeSH-
based profile of the A-literature deemed AP.
The ranked set of weighted index terms is then
filtered leading to a list of selected terms, deemed B-
terms. Filtering consists on selecting important
terms/concepts regarding (1) a specific user selected
topic (Gordon and Dumais, 1998), (2) a user-
selected UMLS semantic type (Weeber et al., 2001;
Srinivasan, 2004; Hu et al.,2010; Chen et al., 2011)
and/or (3) the weighting score (Gordon and Lindsay,
1996; 1999; Srinivasan, 2004; Hu et al., 2010; Chen
et al., 2011).
The filtered terms, also known as B-terms, are
generally used to query PubMed again and to
retrieve B-literature (Gordon and Lindsay, 1996;
1999; Weeber et al., 2001; Srinivasan, 2004; Hu et
al., 2010; Chen et al, 2011). B-literature is again
processed through the same process as A-literature,
leading to C-Concepts.
2
https://www.nlm.nih.gov/research/umls/
Semantic-basedKnowledgeDiscoveryinBiomedicalLiterature
39
C-concepts are considered as potentially correlated
with A-concepts through the intermediate common
B-concepts. The strength of this correlation is
measured using co-occurrence analysis (Stegmann
and Grohmann, 2003), association rules (Hristovski
et al., 2001) or semantically associated relationships
(Liu et al., 2011).
3.2 Closed LBD Approaches
Closed LBD approaches start with A- and C-
concepts. These concepts are respectively used to
query PubMed and to retrieve the corresponding A-
and C-literatures. These working literatures are then
indexed, and common index terms, the intermediate
B-concepts, identified (Weeber et al., 2001). In
(Srinivasan, 2004), the set of common B-Concepts,
also called B-profile or BP, is build as the lexical
intersection of AP and CP MeSH-based profiles
associated with A- and C-literatures respectively.
In this paper, we propose a novel closed LBD
approach that relies on concepts’ semantic
relatedness to identifying intermediate related B-
Concepts from two starting A- and C-literatures. To
evaluate our approach, we use it to replicate the
Swanson’s discovery (Swanson, 1986b) on fish oil
and Raynaud disease connection.
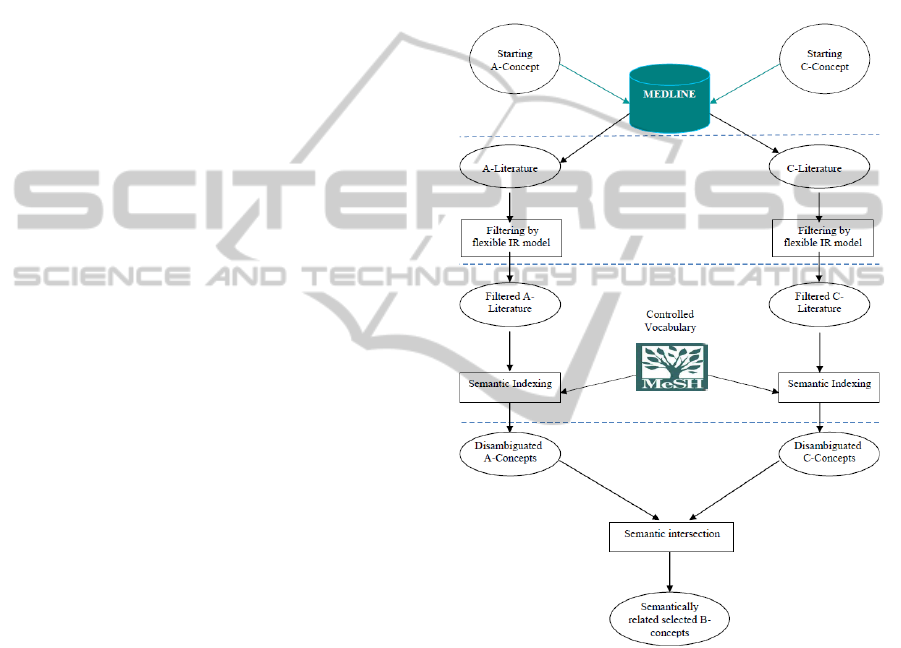
4 PROPOSED APPROACH
In this paper, we propose a semantic-based approach
for closed discovery in the published biomedical
scientific literature. The approach aims at
discovering hidden connections between two starting
concepts of interest, based on the semantic
intersection of the related literatures. For this aim,
our approach relies on: (1) a flexible IR model to
select the most relevant documents from the
MEDLINE retrieved biomedical literature, (2)
MeSH-concepts based indexing to represent these
documents with only representative items, and (3)
concepts’ semantic relatedness (or semantic
similarity) to identify the semantic intersection
between the literatures of interest .
Intuition behind our proposition can be
summarized as follows:
Firstly, using a flexible IR model allows
retrieving a set of ranked documents likely to be
relevant to the given concept of interest, from
which the related literature to use in the
discovery process can be effectively selected
among the highly ranked documents.
Thereafter, indexing with biomedical MeSH
concepts results in better sources of evidence
(intermediate concepts) that will be used for
potential hypothesis validation.
Finally, the semantic intersection of the
literatures of interest allows identifying the
semantically related B-concepts (which may not
be discovered through a lexical intersection).
Our four-step approach is depicted in Figure 3. The
corresponding steps are detailed in the following.
Figure 3: The proposed approach.
4.1 Retrieving A- and C-Literatures
from MEDLINE
Starting from a given hypothesis about a potential
correlation between two focuses (concepts) A and C,
we first have to query PubMed with concepts A and
C respectively. The set of biomedical documents
retrieved by PubMed in response to the query on
concept A- (respectively C-) is the A-literature
(respectively the C-literature).
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
40

4.2 Filtering a- and C-Literatures
through a Flexible IR Model
Filtering a literature consists on selecting its most
relevant documents. For this aim, the concerned
literature is passed through a flexible IR system.
Practically, this consists on using this literature as a
corpus in the flexible retrieval process, and then
querying the flexible IR system based on the related
concept. The retrieved documents are ranked
according to their relevance to the query. Filtering
this retrieved documents consists on keeping the
only most relevant ones (in the following, we limit
the filtered results to the top 1000 relevant
documents for each query). This process is applied
to filter both A- and C-literatures.
4.3 Indexing the Filtered Literatures
with MeSH Concepts
The main objective of this step is to identify the set
of representative biomedical MeSH concepts from
each filtered literature. For this aim, each document
in the filtered literature is indexed, and related
biomedical MeSH concepts identified.
Indexing consists on first extracting
representative terms from the document based on
linguistic text analysis (tokenization, lemmatization,
stop-words elimination), then mapping these terms
onto MeSH entries in order to identify the
corresponding concepts. An ambiguous (polysemic)
term may correspond to several entries (ie. concepts)
in MeSH, it must be disambiguated. To
disambiguate a term, we rely on the disambiguation
approach proposed in (Baziz et al., 2005) for its
simplicity. The underlying idea is to estimate the
semantic relatedness of each MeSH concept with
other MeSH concepts associated with the other
terms of the document. The MeSH concept which
maximizes this estimation is then retained as the
appropriate concept in the indexed document.
The result of this step is a list of representative
disambiguated MeSH concepts associated with each
filtered literature. We denote by Ca the list of A-
concepts associated with A-literature, and by Cc the
list of C-concepts associated with C-literature.
4.4 Identifying Semantically Related
B-Concepts
The objective of this step is to identify the Cb list of
semantically related intermediate B-concepts using
the so-called semantic intersection between Ca and
Cc lists.
To perform semantic intersection between A- and C-
literatures, we rely on the following steps:
Step 1: identifying semantically related concepts
in the two literatures of interest. For this aim, we
propose to calculate the semantic similarity of each
concept C
i
in the Ca list with each concept C
j
in the
Cc list using a semantic similarity measure.
Formally:
Score (C
i
) = Sim(C
i
, C
j
) (1)
Where Sim(C
i
,C
j
) estimates the semantic
similarity between the two concepts C
i
and C
j
on the
basis of the Wu-Palmer measure (Wu et al., 1994).
Highly related concepts, similarity score of
which is higher than a fixed threshold α, in both
literatures are then retained to be examined in the
next step.
Step 2: building the semantic intersection set.
The semantic intersection set is composed of:
Common B-concepts to A- and C-literatures;
Semantically related B-concepts in A- and C-
literatures. In this case, only the is-a semantic
relation is considered. If two concepts X and Y
respectively of A and C-literatures are related
through is-a relation, the most specific concept is
semantically seen as pertaining to both A- and C-
literatures (because if X is-a Y then the specific
concept X is included in Y and not vice versa)
and therefore is included into their semantic
intersection set. For example, if the concept
thrombosis pertains to A-literature and venous
thrombosis pertains to C-literature, knowing that
venous thrombosis is-a thrombosis, then the
venous thrombosis specific concept will be
included in the semantic intersection set of A-
and C-literatures.
5 TESTS AND RESULTS
5.1 Evaluation Settings
We evaluate the performances of our approach by
using it to replicate the Swanson’s first hypothesis
on fish oil and Raynaud disease connection. To
comply with the bibliographic context in which
Swanson made his first discovery:
only the bibliographical references prior to
November 1985 are retrieved from MEDLINE;
only the titles of the bibliographical references
are used in the discovery process;
A-concept is «fish oil», C-concept C is «Raynaud
disease».
To initially retrieve A- and C-literatures from
Semantic-basedKnowledgeDiscoveryinBiomedicalLiterature
41
MEDLINE, we have used the following
keywords_based flexible queries: QA: Fish oil and
QC: Raynaud disease. Then to filter these literatures
through a flexible retrieval model, we used the more
strict concept-based queries: Q’A: “Fish oil” and
Q’C: “Raynaud disease”. These queries allow
retrieving selected literatures of which documents
only contain the considered concepts “Fish oil” (in
case of Q’A) and “Raynaud disease” (in case of
Q’C).
In our experiments, we have tested various
flexible IR models in the filtering step, ranging from
traditional models like Term Frequency Inverse
Document Frequency (TF-IDF) model, probabilistic
models such as OKAPI-BM25 (
Robertson et al., 1998)
and a group of DFR (Divergence from Randomness)
models (
Amati, 2003), as well as language models
(
Ponte et al., 1998). The list of all tested models is
summarized in Table 4.
Table 4: Flexible IR models.
IR Models Description
TF_IDF The tf*idf weighting function.
BM25 The OKAPI probabilistic model.
Dirichlet_L
M
Dirichlet language model.
Hiemstra_L
M
Hiemstra's language model.
DFR_BM25
(DFR)
The DFR version of BM25.
IFB2 (DFR) Inverse term frequency model with Bernoulli’s
process and 2-normalisation for term frequency.
In_expB2
(DFR)
Inverse expected document frequency model for
randomness, the ratio of two Bernoulli's processes
for first normalisation, and 2-normalisation for
term frequency.
In_expC2
(DFR)
Inverse expected document frequency model for
randomness, the ratio of two Bernoulli's processes
for first normalisation, and 2-normalisation for
term frequency with natural logarithm.
InL2 (DFR) Inverse document frequency model for
randomness, Laplace succession for first
normalisation, and 2-normalisation for term
frequency.
LGD (DFR) A log-logistic DFR model.
PL2 (DFR) Poisson model with Laplace succession for first
normalisation, and 2-normalisation for term
frequency.
BB2 (DFR) Bose-Einstein model with Bernoulli's processes for
first normalisation, and 2-normalisation for term
frequency.
All these models are implemented in the generic
Information Retrieval platform IR-ToolKit
3
that we
3
http://www.irit.fr/~Duy.Dinh/tools/irtoolkit/
used for the purpose of our experiments.
Moreover, to extract MeSH concepts from the A-
and C-literatures, we used the Extractor
4
plateform
which implements the state-of-the-art concept
extraction methods.
5.2 Evaluation Protocol
PubMed/MEDLINE is first queried with the related
queries QA and QC respectively, then A- and C-
literatures are retrieved and filtered through a
flexible IR model. The filtered literatures are finally
indexed with MeSH concepts. Semantic relatedness
of each concept in A-literature to each concept in C-
literature is then computed using the Wu-Palmer
similarity score (Wu et al., 1994), and highly related
concepts (the score of which is higher than the fixed
threshold α=0,5) are examined for inclusion in the
semantic intersection set.
The results obtained with semantic intersection
approach are compared to those of a classical lexical
intersection approach performed on PubMed results.
The comparison is mainly performed through the
number of identified concepts (depicted as #
concepts in the following tables), especially at the
level of:
Intermediate B-concepts,
Intermediate B-concepts that are related to
physiology. In this case, we have to check that
blood viscosity and Platelet Aggregation
(identified in early Swanson’s discovery
(Swanson, 1986b)) are among the returned B-
concepts.
5.3 Experimental Results
When asking PubMed/MEDLINE with query QA,
we downloaded a collection of 1092 titles
representing A-literature. When asking it with query
QC, we downloaded a collection of 2692 titles
which represents C-literature. Filtering these
literatures through a flexible IR model leads to 131
(376 for language models) titles representing A-
literature and 380 titles representing C-literature.
The indexing results obtained after filtering A-
and C-literatures and the results of PubMed, as well
as the results of both semantic intersection and
lexical intersection are summarized in Table 5. In
each case, we ensured that B-concepts include blood
viscosity and Platelet Aggregation.
4
http://www.irit.fr/~Duy.Dinh/tools/cxtractor/
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
42
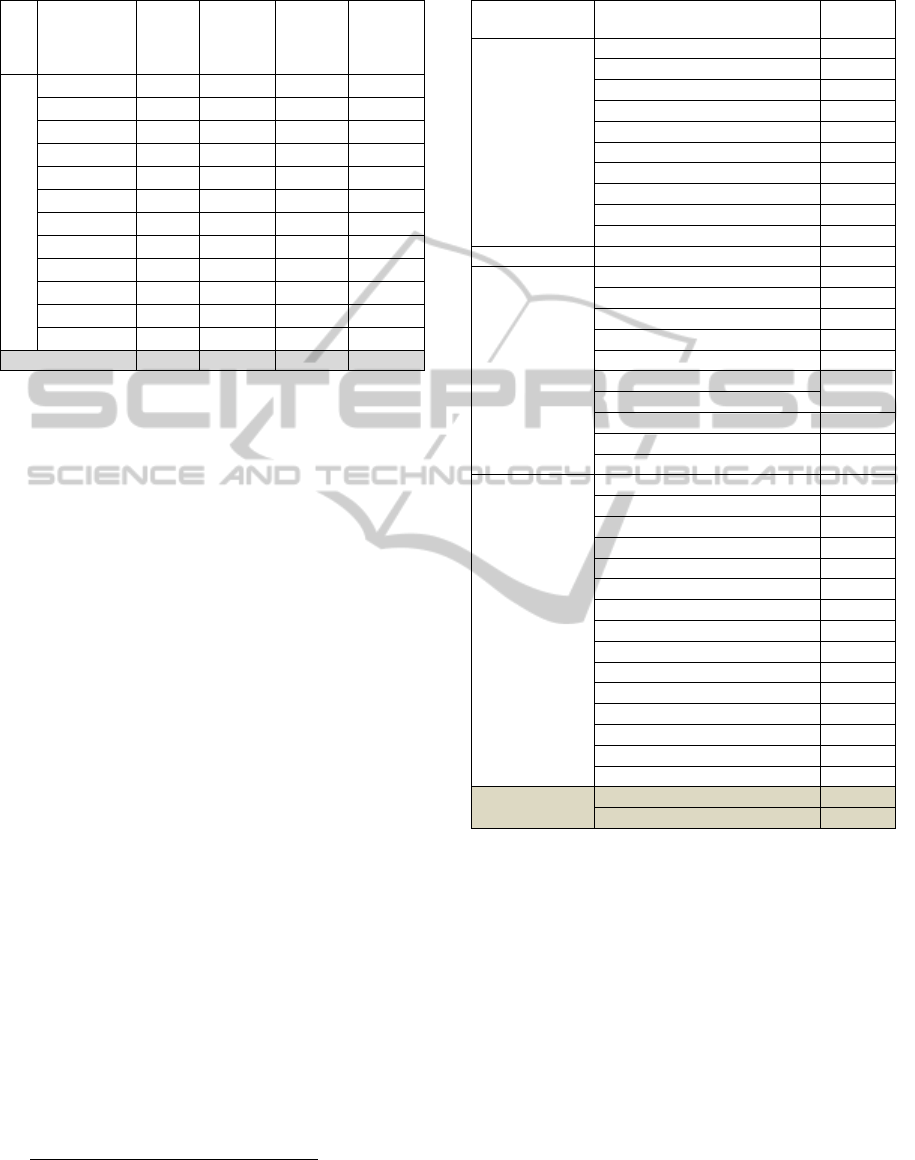
Table 5: Discovered B-concepts.
#
A-
concepts
#
C-
concepts
#
Common
B-
concepts
#
Related
B-concepts
TF_IDF
196 237 32
34
BM25
196 237 32
34
Dirichlet_LM
196 219 31
33
Hiemstra_LM
196 229 32
35
DFR_BM25
196 237 32
34
IFB2
196 237 32
34
In_expB2
196 129 32
34
In_expC2
196 237 32
34
InL2
196 237 32
34
LGD
196 237 32
34
PL2
196 237 32
34
BB2
196 237 32
34
PubMed
236 1052 103 84
The semantic intersection allowed us to identify,
besides the A- and C-literatures common B-
concepts, more than thirty supplementary
semantically related B-concepts that were not
discovered before. Table 6 depicts the B-concepts
that were selected through the semantic intersection
between A- and C-literatures. These concepts are
classified according to their MeSH respective
categories.
When examining the semantic intersection set for
physiology-related (Phenomena and Processes [G]
category) B-concepts, we found the concepts
vasodilation and vasoconstriction that has not been
previously published in Swanson’s works (1986a;
1986b). Whereas vasodilation was discovered by
M.Weeber (Weeber, 2001) using UMLS types and
Metamap
5
(a program to discover UMLS biomedical
concepts referred to in texts), vasoconstriction was
discovered by P. Srinivasan (Srinivasan, 2004)
through his MeSH-profil based approach. To the
best of our knowledge, none of the state-of-art LBD
approaches have discovered these two concepts
simultaneously.
From this point of view, our approach seems to
be promising. This is especially true as we
discovered many other semantically related concepts
of which the ones related to diseases (such as:
venous thrombosis, cerebral infarction, brain
ischemia, …) or to drugs (such as: stanozolol,
epinephrine, apoproteins, …) seem to be interesting
features for assessing new correlations between
Raynaud disease and fish oil and have to be
validated by experts.
5
http://metamap.nlm.nih.gov/
Table 6: Semantically selected B-concepts.
Category B-Concepts Similarity
Score
Anatomy[A]
Aorta 0.89
erythrocyte membrane 0.89
Neutrophils 0.8
Blood platelets 0.75
blood vessels 0.67
bone and bones 0.55
Blindness 0.55
adipose tissue 0.5
Aorta 0.89
erythrocyte membrane 0.89
Organisms [B] Blindness 0.55
Diseases[C]
Hypertriglyceridemia 0.92
Keratoconjunctivitis 0.89
cerebral infarction 0.86
Brain ischemia 0.83
angina pectoris 0.8
thrombosis 0.75
0.67
myocarditis
formates 0.67
syphilis 0.55
conjunctivitis 0.5
Chemicals and
Drugs [D]
dinoprostone 0.88
bilirubin 0.75
vitamin a 0.75
alkaline phosphatase 0.71
norepinephrine 0.67
Nitroglycerin 0.6
Apoproteins 0.57
fish proteins 0.57
polyvinyl chloride 0.57
Carnitine 0.55
stanozolol 0.55
Captopril 0.5
Epinephrine 0.5
cholinesterases 0.5
ethylestrenol 0.5
Phenomena and
Processes [G]
vasodilation 0.55
vasoconstriction 0.55
6 CONCLUSIONS
In this paper, we presented a new semantic-based
approach for biomedical knowledge discovery that
relies on semantic information retrieval techniques.
This approach brings a novel method for
discovering unknown correlations between
biomedical concepts through the so-called semantic
intersection between the corresponding semantically
indexed literatures. Using this approach to replicate
Swanson’s early discovery leads to discovering new
B-concepts that were not previously identified in the
state-of-art LBD works. This seems a very
promising issue for LBD.
Flexible Retrieval Models
Semantic-basedKnowledgeDiscoveryinBiomedicalLiterature
43

Works are in progress to first validate the newly
discovered B-Concepts, and then to experiment the
proposed approach on an open discovery process.
REFERENCES
Amati G., 2003. Probabilistic models for Information
Retrieval based on Divergence from Randomness,
PhD Thesis, University of Glasgow.
Baziz M., Boughanem M., Aussenac-Gilles N., 2005. A
Conceptual Indexing Approach for the TREC Robust
Task. In The Fourteenth Text REtrieval Conference
Proceedings (TREC 2005), Gaithersburg, Maryland,
15/11/2005-18/11/2005. E. M. Voorhees, Lori P.
Buckland (Eds.), NIST, November 2005.
Chen R., Lin H., Yang Z., 2011. Passage retrieval based
hidden knowledge discovery from biomedical
literature. Expert Systems with Applications, pp. 9958–
9964.
Coletti M. H., and Bleich H. L., 2001. Medical subject
headings used to search the biomedical literature.
Journal of the American Medical Informatics
Association, Vol. 8, pp. 317–323.
Ganiz M. C., Pottenger W. M., Janneck C. D, 2005.
Recent Advances in Literature Based Discovery.
Journal of the American Society for Information
Science and Technology, JASIS.
Gordon M. D., Lindsay R. K., 1996. Towards discovery
support systems: a replication, re-examination, and
extension of Swanson’s work on literature-based
discovery of a connection between Raynaud’s and fish
oil. Journal of the American Society for Information
Science, Vol. 47, p.116-128.
Gordon M. D., Dumais S., 1998. Using Latent Semantic
Indexing for literature based discovery. Journal of the
American Society for Information Science and
Technology, Vol. 49, p. 674-685.
Gordon M. D., Lindsay R. K., 1999. Literature-based
discovery by lexical statistics. Journal of the American
Society for Information Science, Vol. 50, p.574-587.
Hristovski D., Stare J., Peterlin B., Dzeroski S., 2001.
Supporting discovery in medicine by association rule
mining in MEDLINE and UMLS. Medinfo, Vol. 10, p.
1344-1348.
Hu X., Zhang X., Yoo I., Zhou X., Xu X., 2010. Mining
Hidden Connections among Biomedical Concepts
from Disjoint Biomedical Literature Sets through
Semantic-Based Association Rule. International
Journal of Intelligent Systems, Vol. 25, Issue 2,
(February 2010), pp. 207-223.
Liu H., Le Pendu P., Ruoming Jin, Dejing Dou., 2011. A
Hypergraph-based Method for Discovering
Semantically Associated Itemsets. In Proceedings of
the 2011 IEEE 11th International Conference on Data
Mining (ICDM '11). IEEE Computer Society,
Washington, DC, USA, pp. 398-406.
Liu X., Fu H., 2012. Literature-based knowledge
discovery: the stat of the art. CoRR abs/1203.3611.
Ponte J. M., Croft W. B. A Language Modeling Approach
to Information Retrieval. In Proceedings of the 21st
annual international ACM SIGIR conference on
Research and development in information retrieval,
pp. 275-281
Robertson S. E., Walker S., Hancock-Beaulieu M., 1998.
Okapi at TREC-7: Automatic AdHoc, Filtering, VLC
and Interactive. In Proceedings Text REtrieval
Conference, TREC-7, p.199–210.
Srinivasan P., 2004. Text Mining Generating Hypotheses
from MEDLINE. Journal of the American Society for
Information Science and Technology, Vol. 55, pp.396-
413.
Stegmann J., Grohmann G., 2003. Hypothesis generation
guided by co-word clustering. Scientometrics, Vol. 56
N°1, pp. 111–135.
Swanson D. R., 1986a. Undiscovered public knowledge.
Library Quarterly, Vol. 56, N°2, pp. 103-118.
Swanson D. R., 1986b. Fish oil, Raynaud’s syndrome, and
undiscovered public knowledge. Perspectives in
Biology and Medicine, Vol 30, p.7-18.
Swanson D. R., 1988. Migraine and magnesium: eleven
neglected connections. Perspectives in Biology and
Medicine, Vol 31, pp. 526-557.
Swanson D. R., 1989. Online search for logically-related
noninteractive medical literature: A systematic trial-
and-error strategy. Journal of the American Society of
Information Science, Vol. 40, pp. 356-358.
Weeber M., Klein H., de Jong-van den Berg L. T. W. ,
2001. Using concepts in literature-based discovery:
Simulating Swanson’s Raynaud – fish oil and
Migraine – magnesium discoveries. Journal of the
American Society for Information Science and
Technology, Vol. 52, pp. 548-557.
Wu Z., Palmer M., 1994.Verb semantics and Lexical
selection. In Proceedings of the 32th Annual Meetings
of the Association for Computational Linguistics, pp.
133-138.
KDIR2013-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
44
