
BIOscrabble
Extraction of Biological Analogies out of Large Text Sources
Maria Katharina Kaiser, Helena Hashemi Farzaneh and Udo Lindemann
Institute of Product Development, Mechanical Engineering, Technische Universität München,
Bolzmannstrasse 15, 85748 Garching, Germany
1 STAGE OF THE RESEARCH
The aim of the research presented here is a support
for solution search in bio-inspired design. More
specifically, the engineer shall be supported in
extracting biological analogies out of large text
sources whereby the biological analogies must have
the potential to solve given technical problems. To
reach this aim, an approach has been developed by
Kaiser (Kaiser, 2012) which in the first instance
focuses on
the large text source that shall be scanned and
the search terms that shall be used to describe the
technical problem.
As the proposed text source – biological research
articles – is large and, therefore, the extraction of
useful analogies is challenging, the approach
proposed by Kaiser (Kaiser, 2012) needs further
development. Research started on how to manage
large amounts of information. In contrary to the
research on existing support for bio-inspired design,
research on managing large amounts of information
is still in the early stages. Initial case studies have
been conducted in order to test the applicability and
practicability of the existing approach and to
understand how to support information management
in bio-inspired design.
2 RESEARCH PROBLEM
The voyage of discovery is not in seeking new
landscapes but in having new eyes.
Marcel Proust
The question of how the huge amount of already
recorded biological information can be discovered
and used for developing innovative technical
solutions has been driving engineers for centuries. In
the recent decades, research activities in the field of
bio-inspired design intensified. To effectively
support the exploitation of existing biological
solutions for designing technical products, two main
questions can be addressed: 1) How can a promising
biological solution be identified by an engineer who
does not have a biological background? and 2) How
can this engineer transfer a promising biological
solution into a technical concept or product?
This research focuses on answering the first
question. In literature, two approaches can be
distinguished. On the one hand, databases have been
established to provide the engineer with edited and
often simplified biological knowledge
(http://www.asknature.org/, (Chakrabarti, 2005);
(Löffler, 2008); (Gramann, 2004); (Hill, 1997). On
the other hand, researchers tried to find ways to
support the engineer in effectively using biological
knowledge that is available in text format (research
on this area is discussed in detail in Sec. 5). In
contrast to databases, the latter approaches do not
face challenges such as the need of being initially
filled and kept up-to-date or the risk of accessing
subjectively misinterpreted biological data.
Therefore, the authors decided on providing further
support in this research area.
Searching biological analogies in biological
literature is not trivial: a search term-based search in
these sources can display an enormous amount of
biological information. It is therefore challenging for
an engineer to identify the right biological analogy
for a given technical problem. This is especially true
for searching in biological research articles.
Nevertheless, this search source is very
comprehensive and represents the current state of
biological research and is hence worth further
consideration.
3 OUTLINE OF OBJECTIVES
This paper illustrates an approach – called
BIOscrabble – that aims at supporting solution
search in bio-inspired design or, to be more precise,
at supporting the search term-based extraction of
10
Kaiser M., Hashemi Farzaneh H. and Lindemann U. (2013).
BIOscrabble - Extraction of Biological Analogies out of Large Text Sources.
In Doctoral Consortium, pages 10-20
Copyright
c
SCITEPRESS

relevant biological analogies out of large biological
text sources. The objectives of this paper are:
to present the approach BIOscrabble that
- supports the engineer in performing search
term-based searches in large biological text
sources. The engineer is advised to choose
search terms that describe the technical
problem entirely and aptly.
- proposes prospects for supporting the engineer
in managing a large number of search results
by using a graph-based representation of the
search.
to discuss the implications of the application of
BIOscrabble for the search process and the
organization of the search results.
4 STRUCTURE OF THIS PAPER
In Sec. 5 an overview on related research activities
in the field of discovering biological analogies in
text sources is given. Sec. 6 and 7 illustrate the
BIOscrabble approach and the BIOscrabble software
prototype. The application of BIOscrabble is
discussed in Sec. 8. The discussion is based on
findings from case studies in which BIOscrabble
was applied to technical problems. The expected
outcome of the BIOscrabble research project is
illustrated in Sec. 9. The paper concludes with an
outlook on future work in Sec. 10.
5 STATE OF THE ART
Research activities have been conducted that focus
on the question of how to effectively discover
biological information available in text format. In
the following, an overview over research activities is
given that address the issues of choosing text
sources and search terms, overcoming linguistic
differences between engineers and biologists and
identifying relevant biological analogies.
5.1 Text Sources
Which text sources have been proposed for an
effective discovery of biological information
available in text format so far?
A natural language approach to biomimetic
design has been developed at the University of
Toronto. Keyword searches were performed and
analysed in an introductory biological textbook to
support an effective retrieval of biological
information available in text format. The textbook
was chosen as an initial text source, because 1) it is
intelligible for a reader without a biological
background and 2) it provides biological information
ranging from molecular structures to ecosystems
(Shu, 2010); (Cheong, 2012).
Biological textbooks were also used by Stroble et
al. and Nagel et al. (Stroble, 2009); (Nagel, 2010)
for compiling engineering-to-biology thesauri and
by Nagel and Stone (Nagel, 2011) for testing a
methodology for facilitating systematic biologically
inspired design (including searching for biological
analogies).
These textbook-based approaches can be applied
to any other text source. However, advanced sources
such as biological research articles are suggested for
finding further details on selected biological
analogies rather than for initial searching (Shu,
2010).
The World Wide Web was analysed as an initial
source from which biological inspiration can be
drawn by Vattam and Goel (Vattam, 2011).
Searching the web for relevant biological analogies
was found to be very costly in terms of time spent on
searching and analysing (resource costs). The
probability of spending time on analysing useless
information or of ignoring useful information due to
uncertainty was found to cause additional costs
(opportunity costs). Using the web as an initial
source for searching for biological analogies was
also explored and supported by Vandevenne
(Vandevenne, 2011; 2012b). The approaches are
further described in Sec. 5.4.
The approaches described in Sec. 5.1 did not
focus on using biological research articles as an
initial search source for discovering biological
information available in text format. As biological
research articles are representing biological research
most comprehensively, they are used as an initial
search source in the approach proposed here.
5.2 Search Terms
What types of search terms have been proposed for
an effective discovery of biological information
available in text format so far?
To search biological texts for relevant biological
analogies, at the University of Toronto, primarily
verbs describing the desired technical function or
effect were used as search terms (Shu, 2010);
(Cheong, 2012). In one work the use of adjectives
describing the desired technical qualities was
illustrated (Ke, 2010). Nouns were found to indicate
preconceived solutions and were therefore neglected
BIOscrabble-ExtractionofBiologicalAnalogiesoutofLargeTextSources
11

(Shu, 2004; 2010).
Functional technical terms or terms of the
Functional Basis (widely accepted characterization
of product functions in a verb-object (function-flow)
format (Stone, 2000) serve as basis for the biological
terms in the engineering-to-biology thesauri
developed by Stroble et al. and Nagel et al., (Stroble,
2009); (Nagel, 2010). The engineering functional
terms contained in the thesauri play a central role in
searching for relevant analogies in the methodology
proposed by Nagel and Stone (Nagel, 2011) which
aims at facilitating systematic biologically inspired
design.
For identifying biological analogies, functions or
functional verbs of technical or biological systems
are central in the work of Vandevenne (Vandevenne
2011).
It can be concluded that the approaches
described in Sec. 5.2 mainly focus on system
functions to discover biological information
available in text format. In the approach proposed
here, search terms describing system properties and
environmental influences on systems are
additionally focused on.
5.3 Terminology
What options have been revealed to bridge the gap
between an engineer’s and a biologist’s terminology
so far?
At the University of Toronto, two alternatives
were revealed which can mitigate linguistic
differences between engineers and biologists. First,
it was found that the chance of identifying relevant
biological analogies in biological text sources
increases when the engineering functional search
terms are expanded by term variations such as
synonyms (Vakili, 2001); (Hacco, 2002). Second, a
method for identifying biologically meaningful
keywords was developed (Chiu, 2005; 2007).
Textbook words collocated with the functional
search terms initially used for searching were
defined as biologically meaningful keywords
dependent on their occurrences in further text
sources. Biologically meaningful keywords were
furthermore defined for the terms of the Functional
Basis (Cheong, 2008).
To link engineers’ and biologists’ terminology
and, therefore, sharpen search results when
performing a key word search, engineering-to-
biology thesauri have been developed by other
research groups (Stroble, 2009); (Nagel, 2010).
Biologically meaningful keywords or keywords
contained in the engineering-to-biology thesauri are
only available for a portion of functional search
terms. Therefore, in this work common term
variations such as synonyms are used for varying
search terms.
5.4 Identifying Biological Analogies
What decision guidance has been given for
effectively identifying relevant biological analogies
described in text format so far?
Researchers at the University of Toronto
approached this question as follows: 1) Hacco and
Shu detected characteristics of biological texts that
contain irrelevant biological analogies (Hacco,
2002). When searching for biological analogies,
these characteristics can form the basis for a
systematic removal of irrelevant search results and,
therefore, assist the engineer in identifying relevant
analogies. 2) A characteristic of biological texts that
can effectively be used as design stimuli is the
explicit description of biological principles (Mak,
2004). A case study showed that the effort for
selecting relevant biological systems is reduced
when the principle has not to be abstracted from
described behaviors or forms. 3) To further support
the engineer in identifying relevant analogies,
different methods for categorizing biological
information have been developed based on semantic
relations (Ke, 2009); (Cheong, 2012); (Son, 2012).
Categorized biological information was found to
assist the engineer in selecting analogies relevant to
the design problem (Ke, 2009); (Cheong, 2012).
Further support for identifying relevant
biological analogies was provided by Vandevenne
(Vandevenne, 2011; 2012a; 2012b) in different
ways: 1) To support the identification of relevant
analogies, textual descriptions of biological systems
that are detectable in the web were automatically
characterized and mapped to technical problem
descriptions (Vandevenne, 2011). 2) To support the
structuring and, therefore, the selection of biological
information available in text format, Vandevenne et
al., introduced a method for classifying biological
analogies into the Biomimicry Taxonomy of the
Biomimicry Design Portal Ask Nature
(http://www.asknature.org/, Vandevenne, 2012a). 3)
For filtering out and preselecting biological
information that is available in the web, a
webcrawler was developed that continuously
collects documents containing biological
information that is relevant to biomimetics
(Vandevenne, 2012b).
In the approaches described in Sec. 5.4, no
assistance for identifying biological analogies
IC3K2013-DoctoralConsortium
12

described in biological research articles was
provided explicitly. Nevertheless, concepts such as
the categorisation or clustering of biological
information are used in the approach proposed here.
6 BIOscrabble – APPROACH
As with the approaches described, BIOscrabble
supports the effective discovery of biological
information available in text format. The main
differences to the existing approaches described in
Sec. 5 are: 1) the exclusive use of biological
research articles as an initial search source, 2) the
explicit inclusion of search terms that describe a
desired technical system’s properties and the
environmental influences on this system and 3) the
use of a graph-based representation of the search
that has been carried out for assisting the engineer in
identifying relevant biological analogies.
In the following, the BIOscrabble approach and a
BIOscrabble software prototype are illustrated.
BIOscrabble is a further development of the
approach proposed by Kaiser (Kaiser, 2012). In the
following a short overview over BIOscrabble is
given. Further descriptions of the BIOscrabble steps
and components as well as the reasons for their
inclusions are given in Sec. 6.2 – 6.5.
6.1 Overview
For the effective discovery of biological information
available in text format, the following steps are
proposed in BIOscrabble (Fig. 1):
1) The user – here the engineer – describes the
technical problem under consideration in terms
of system functions, system properties and the
environmental influences on it (definitions see
below). The search terms for the following
search process are taken from this description.
2) The search terms are varied, e.g. synonyms are
built. WordNet (description see below) provides
assistance here.
3) The database PubMed (description see below) is
searched using the original and the varied search
terms.
4) The PubMed search results, i.e. the biological
research articles are clustered and displayed to
the user in the form of structured graphs.
6.2 Text Sources
BIOscrabble supports using biological research
articles as initial text sources when searching for
biological analogies via search terms.
Figure 1: BIOscrabble search process.
BIOscrabble-ExtractionofBiologicalAnalogiesoutofLargeTextSources
13

In BIOscrabble, PubMed is used as a search
source (http://www.ncbi.nlm.nih.gov/pubmed/).
PubMed is a meta-database (i.e. a database of
databases) that refers to more than 22 million
citations for biomedical literature. Besides citations
in the field of biomedicine, health and
bioengineering, life sciences, behavioural sciences
and chemical sciences are addressed. PubMed was
developed and is maintained by the National Center
for Biotechnology Information (NCBI). It can be
freely used. PubMed can be searched by search
terms. Search terms can be connected by Boolean
operators such as AND or OR. Connecting search
terms with AND results in citations that must
contain all search terms connected. Connecting
search terms with OR results in citations that must
contain at least one of the search terms connected.
PubMed provides different filters that can be set
when performing a search. The BIOscrabble user is
advised to set the filter “abstract available”. This
filter secures that at least an article’s abstract is
available in addition to its title. This is the minimum
requirement for the identification of relevant search
results.
Biological research articles were chosen as an
initial search source, because this text source is very
comprehensive and represents the current state of
biological research. In biological textbooks, not all
biological research findings are illustrated or
illustrated in detail. This was stated in interviews
with biologists that were conducted within a project
aiming at the development of a communication
platform for engineers and biologists.
6.3 Search Terms
BIOscrabble advises to search for biological
analogies in biological research articles by search
terms that describe a desired technical system’s
functions, properties or the environmental influences
on this system. This search schema encourages the
engineer to see the considered technical problem or
system from different angles which can increase the
diversity of the search terms. This, in turn, is
supposed to enhance the probability of detecting
relevant biological analogies.
6.3.1 Functions
Search terms describing technical functions are
included in BIOscrabble because of two reasons: 1)
Existing research dealing with the discovery of
biological analogies in biological text sources
showed that good results can be obtained by using
functional search terms (Shu, 2010, Vandevenne,
2011, Cheong, 2012). 2) Functions play a key role in
modelling technical systems (Stone, 2000, Pahl,
2007, Erden, 2008). Engineers are therefore trained
in describing a technical system by its functions.
In this work, functions are defined according to
Pahl et al., (2007).
6.3.2 Properties
BIOscrabble includes search terms describing
technical system properties for the following reason:
Compared with engineers, biologists do not focus on
describing biological systems in terms of functions
as the concept of function is not an issue during their
academic training. Therefore, biological systems
which are not primarily described functionally can
be missed when searching with functional search
terms only. In a pre-study carried out by the authors,
spider silk, for example, was found to be described
in terms of system properties rather than system
functions.
Here, properties are defined according to Eder
qnd Hosnedl (2008).
6.3.3 Environmental Influences
Natural systems are strongly aligned to the
environmental influences they are exposed to as they
evolve driven by their environment. Therefore,
search terms that describe the environmental
influences on a technical system are assumed to lead
to relevant biological analogies. The above named
pre-study confirmed this assumption. Consequently,
environmental influences are included in
BIOscrabble as a search term type.
In this work, the environment is defined
according to Srinivasan and Chakrabarti (2009).
Environmental influences are hence the influences
of the environment on a technical system.
6.4 Terminology
To account for differences between an engineer’s
and a biologist’s terminology, the BIOscrabble user
is advised to vary the search terms (original search
term) he or she picks. The following search term
variations (varied search term), some of which are
supported by WordNet (see below), are included in
BIOscrabble: 1) synonyms of the search term, 2)
nouns, verbs and adjectives derived from the search
term and 3) antonyms of the search term. If the
search term would be “purifying”, 1) could be
“cleansing”, 2) would be “purification, purify and
pure” and 3) would could be “adulterating”.
IC3K2013-DoctoralConsortium
14

Synonyms are included as they were found to
increase the chances of getting relevant matches in
case of searching a biological textbook for biological
analogies via engineering functional search terms
(Vakili, 2001); (Hacco, 2002). The authors assume
that this is also true for searching biological research
articles with search terms describing technical
functions, properties or environmental influences.
Deriving nouns, verbs and adjectives from a search
term has the potential to broaden the solution space
without leading to results that vary from the topic.
BIOscrabble uses WordNet for automating some of
the above mentioned search term variations
(http://wordnet.princeton.edu/). WordNet is a lexical
database of English nouns, verbs, adjectives and
adverbs which are related by cognitive synonymy.
WordNet was also used at the University of Toronto
(Hacco, 2002); (Chiu, 2007); (Ke, 2010); (Cheong,
2012) for varying the terms used for searching
biological text sources for analogies.
6.5 Identifying Biological Analogies
BIOscrabble proposes to use graph-based
representations of search processes in bio-inspired
design to support the engineer in identifying
biological analogies that are hidden in large text
sources.
Graphs are abstract structures which represent
objects and their relations. Objects are represented
as nodes, relations are represented as edges. Here,
typed attributed graphs are used that contain three
kinds of node types and two kinds of edge types: the
node types are 1) original search term, 2) varied
search term and 3) research article, the edge types
are 1) edges linking search terms to their variations
and 2) edges linking search terms (original or
varied) to the research articles in which they are
contained.
Graphs showing the relations between the
original search terms and the varied search terms,
between the search terms (original and varied) and
the corresponding research articles or between
different research articles can support the
identification of biological analogies in different
ways. The engineer can work with graphs that
cluster research articles in which the same search
terms are contained
cluster research articles in which the same types
of search terms (function, property,
environmental influence; original term, varied
term) are contained
cluster research articles in which the same or
different search terms are contained with a
certain frequency
cluster research articles which are cited by each
other
cluster research articles which are written by the
same first authors
These clusters can be supportive. Research article
samples can be taken within each cluster and
examined for relevant biological analogies. If the
samples within one cluster are promising the cluster
can be examined further. The large amount of
research articles a PubMed search generally
produces therefore is divided into manageable
“packages”. The benefit of categorizing or clustering
biological information is shown in (Ke, 2009,
Vandevenne, 2011, 2012a, Cheong, 2012).
7 BIOscrabble – SOFTWARE
PROTOTYPE
From a certain amount of search results (> 50)
onwards it is impracticable to generate these graph
views manually. Therefore, the BIOscrabble search
process including the generation of the search graph
has been automated in a software prototype.
7.1 Front End
By default the user is asked to enter search terms
describing system functions, system properties and
environmental influences on the desired system.
These different types of search terms can be
connected via check marks in order to search for
research articles that contain more than one type of
search terms. Check marks correspond to the
Boolean operator AND. Empty check boxes
correspond to the Boolean operator OR. In this case
all research articles are searched in which at least
one type of search terms is contained. To exclude
research articles containing particular terms,
unwanted terms can be added.
The user has the possibility to exclusively search
for review articles by setting the filter “Review
Articles”. Research articles which are freely
available in the full text version can be searched for
by setting the filter “Fulltext Available Articles”.
As a result of the search process, a table is
displayed. This table contains the titles and the first
authors of the resulting research articles as well as
their rank and their rating. The rank increases with
the value of the rating. Both are dependent on the
BIOscrabble-ExtractionofBiologicalAnalogiesoutofLargeTextSources
15
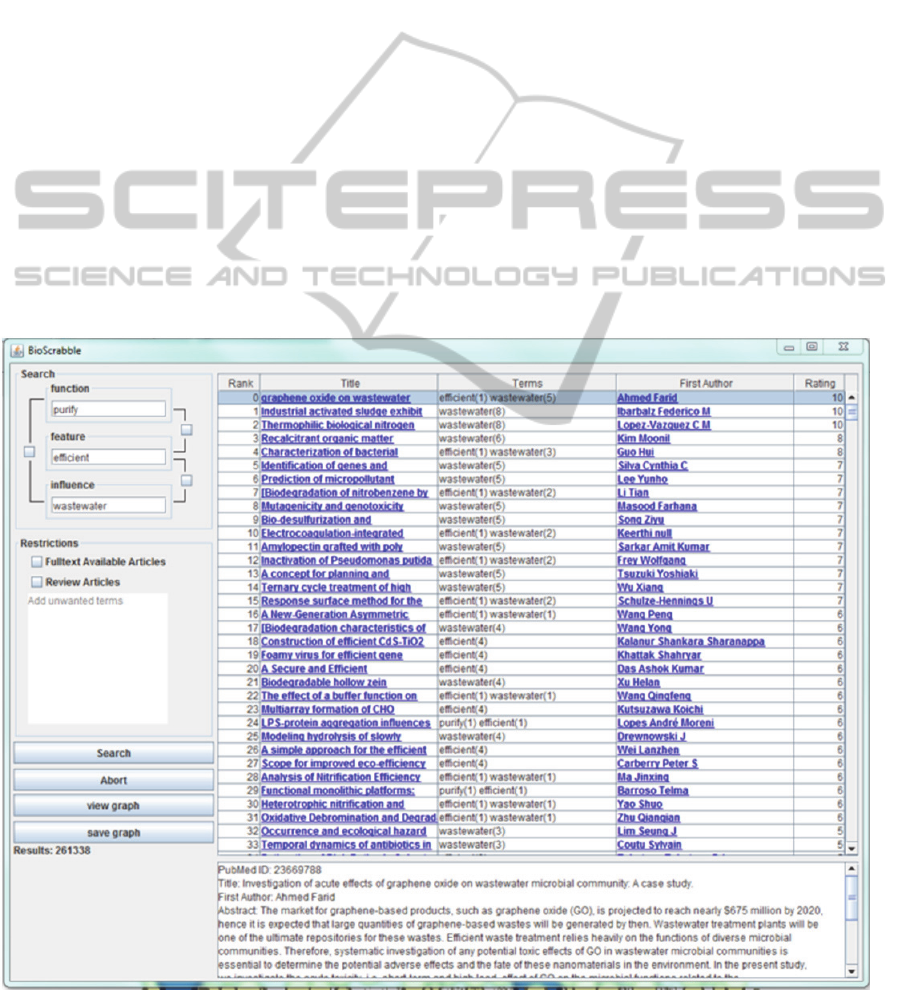
search terms’ frequency in each search result (see
7.2).
The search results can be sorted by their rank,
their title, their terms, their first authors and their
rating.
For every search process the corresponding
search graph can be viewed and saved as well as
exported as a .graphml file. In the graph, information
about the node and edge types as well as information
about the rank and rating of the resulting research
articles is contained.
Fig. 2 shows the graphical user interface (GUI)
of BIOscrabble.
7.2 Back End
After the search process has been started by the user,
the search terms which have been entered are varied.
For the variation of the search terms WordNet
Search – 3.1 is used online. Each search term is
connected to its variations by the Boolean operator
OR.
Depending on the user input, the different types
of search terms are connected by either the Boolean
operator AND or OR. The resulting search phrase is
used to search PubMed for biological research
articles. The PubMed filter “abstract available” is set
automatically for the reasons mentioned in Sec. 6.2.
The search terms (original and varied) that are
contained in an article’s title or abstract are counted.
For the generation of the search graph the
following information is extracted from the search
process: 1) which term is the origin or the variation
of which other term, 2) which term is contained in
which article and 3) how frequently is a term
contained in one article (necessary for representing
the article’s rank and rating).
With the current software prototype it is possible
to distinguish articles according to the frequency of
the search terms that are contained. As explained in
Sec. 7.1 the frequency of the search terms is
reflected in the rating and rank of the articles. The
rating is calculated as shown in equation 1:
rating = 2 * a + b (1)
In equation 1, a is the number of different search
terms contained in a research article and b is the
total number of search terms found in the article.
Figure 2: Screenshot of BIOscrabble GUI.
IC3K2013-DoctoralConsortium
16

Equation 1 reflects one of the observations made
during the application of the BIOscrabble approach
to different technical problems in case studies with
students of mechanical engineering (see 8.3).
Fig. 3 shows two graphs of an exemplary search.
The left graph shows the raw representation of the
relations between the search terms and the research
articles they are contained in. The right graph shows
the same relations, but the rank of the articles is
reflected in the size of the nodes representing them,
i.e. articles with a higher frequency of search terms
are represented in bigger nodes.
The implementation of other graph-based
representations (see above) is in the concept stage.
8 DISCUSSION
In the following, the approach of BIOscrabble is
discussed based on the documentation of case
studies in which BIOscrabble was applied to
different technical problems. In these case studies,
students of mechanical engineering were asked to
solve a given technical problem applying to the
search process proposed by BIOscrabble and to
document their results. They did not use the software
prototype and, therefore, were not supported in the
identification of biological analogies. For brevity,
the technical problems are not discussed in detail
here.
The proposed text source, the proposed types of
search terms including the proposed variations and
the proposed support for the identification of
biological analogies are discussed separately.
8.1 Text Sources
Biological research articles which are available via
PubMed proved to be a text source which can
provide useful biological information for the
developments of bio-inspired products. Besides
articles about biological systems, articles that
describe bio-inspired or other technical products
which are used in a biological context were found to
be useful for the solving of the considered technical
problems.
Figure 3: Exemplary graphs; left: raw representation, right: edited representation; black: node type original search term,
grey: node type varied search term, white: node type research article.
BIOscrabble-ExtractionofBiologicalAnalogiesoutofLargeTextSources
17

Therefore, PubMed can be viewed as a search source
that is not only useful for the development of bio-
inspired products, but can also serve as a tool for
finding analogies in general.
For most PubMed searches an impracticable
amount of research articles is displayed. Without
additional support it is very time-consuming to
identify articles that contain useful biological
information. To make a high amount of search
results manageable within a reasonable time, graphs,
as described above, can be supportive.
In conclusion, PubMed is a promising text source
for finding biological or other analogies for solving
technical problems. However, the engineer has to be
supported in exploring the huge amount of search
results. Furthermore, biological research articles are
more appropriate for getting inspiration of what
biological systems exist than for understanding the
basic principles behind them. For engineers, many of
them are hard to understand.
8.2 Search Terms
The case studies show that depending on the
technical problem different types of search terms led
to useful biological analogies.
In most cases, the best results were obtained by
searching with AND combinations of search terms
of the types function, property and environmental
influence. Nevertheless, there were technical
problems that were solved using biological systems
contained in articles that were found by only one
type of search term. Examples are the design of an
adaptive surface with variable heat conductivity, a
self-sharpening knife or a tension-reducing
mechanism for stuffed pieces of luggage. In case of
the adaptive surface, functional search terms were
most beneficial. For the knife and the tension-
reducing mechanism, search terms of the type
system property and environmental influence led to
the best analogies.
Search term variations in all cases led to
additional relevant biological analogies.
It is concluded that the types of search terms that
are proposed as well as the search term variations
are beneficial for the discovery of biological
analogies in large text sources. Whether the benefit
of certain search term types or term variations
depends on the kind of technical problem remains to
be proven.
8.3 Identifying Biological Analogies
The graph-based representations proposed in Sec.
6.5 were not available for the participants of the case
studies as they did not use the BIOscrabble software
prototype. Nevertheless, conclusions concerning a
support for identifying biological analogies can be
drawn from the relations between the search terms
that were used for searching PubMed and those
research articles that contain relevant biological
analogies.
For the development of a device for purifying
drinking water, the relevance of the research articles
correlated with the frequency of search terms that
are contained. Additionally, a slight correlation
between the relevance of an article and the
frequency of different types of search terms was
observed. These findings are realized in the paper
rating and ranking mechanism that is implemented
in the current BIOscrabble software prototype (see
Sec. 7).
For the other design case studies, this
observation did not prove true. In case of the self-
sharpening knife the most relevant research articles
contained only one single search term. Although, all
articles that were relevant for the development of a
self-sharpening knife dealt with only two topics,
namely “teeth” and “claws”. Therefore, a graph that
shows clusters of research articles that are cited by
each other or are written by the same first author –
and thus are supposed to deal with a similar topic –
is proposed.
Whether the other graphs that are proposed in
Sec. 6.5 can be beneficial remains to be proven.
9 EXPECTED OUTCOME
The expected outcome of this research project is to
provide a functional software tool for supporting
solution search in bio-inspired design. The expected
partial outcomes are the following:
BIOscrabble will provide guidelines
- supporting the formulation of promising search
terms for finding biological analogies for a
technical problem. Depending on the technical
problem, guidelines are given which search
term type is most beneficial for searching.
- supporting the selection of term variations
according to their potential of increasing the
amount of relevant analogies.
- supporting a type of technical problem-specific
use of the graph-based representations.
BIOscrabble will provide the possibility to
execute graph-based analyses in order support
the user in exploring the huge amount of
biological information.
IC3K2013-DoctoralConsortium
18

The above mentioned partial outcomes will be
based on case studies in which BIOscrabble is
applied to a sufficiently large number of different
classes of technical problems.
BIOscrabble will be connected to a platform
supporting the transfer of biological solutions
into technical solutions that is developed in
another research project.
10 FUTURE WORK
To achieve those outcomes, further research has to
be done and further case studies with the application
of BIOscrabble have to be performed to
understand possible correlations between types
of search terms and relevant biological research
articles dependent on specific classes of technical
problems.
understand possible correlations between the use
of search term variations and relevant biological
research articles dependent on specific classes of
technical problems.
understand possible correlations between the
benefit of certain graphs and specific classes of
technical problems.
extend the graphs that are currently proposed.
to further prove the text source of biological
research articles (and the database PubMed), the
types of search terms, the term variations (and
the software WordNet) and the graphs regarding
their value for solution search in bio-inspired
design.
ACKNOWLEDGEMENTS
The authors want to thank the students who applied
the BIOscrabble approach in their biomimetic
development projects as well as the students who
added to the development of the BIOscrabble
software prototype.
REFERENCES
Chakrabarti A., Sarkar P., Leelavathamma B., and
Nataraju B. S., 2005, A Functional Representation for
Aiding Biomimetic and Artificial Inspiration of New
Ideas, Artificial Intelligence for Engineering Design,
Analysis and Manufacturing, 19(02), pp. 113–132.
Cheong H., Shu L. H., Stone R. B., and McAdams D. A.,
2008, Translating Terms of the Functional Basis into
Biologically Meaningful Keywords, Proceedings of
IDETC/CIE 2008 ASME 2008 International Design
Engineering Technical Conference & Computers and
Information in Engineering Conference, New York
City, NY, USA.
Cheong H., and Shu L. H., 2012, Automatic Extraction of
Causally Related Functions from Natural-Language
Text For Biomimetic Design, Proceedings of the
ASME 2012 International Design Engineering
Technical Conferences & Computers and Information
in Engineering Conference IDETC/CIE 2012,
Chicago, IL, USA.
Chiu I., and Shu L. H., 2005, Bridging Cross-Domain
Terminology for Biomimetic Design, Proceedings of
IDETC’05 ASME 2005 International Design
Engineering Technical Conferences and Computers
and Information in Engineering Conference, Long
Beach, California, USA.
Chiu I., and Shu L. H., 2007, Biomimetic Design through
Natural Language Analysis to Facilitate Cross-Domain
Information Retrieval, Artificial Intelligence for
Engineering Design, Analysis and Manufacturing,
21(01), pp. 45–59.
Eder W. E., and Hosnedl S., 2008, Design-Engineering –
A Manual for Enhanced Creativity, Taylor & Francis
Group LLC.
Erden M. S., Komoto H., Van Beek T. J., D’Amelio V.,
Echavarria E., and Tomiyama T., 2008, A review of
function modeling: Approaches and applications,
Artificial Intelligence for Engineering Design,
Analysis and Manufacturing, 22(02), pp. 147–169.
Gramann J., 2004, Problemmodelle und Bionik als
Methode.
Hacco E., and Shu L. H., 2002, Biomimetic Concept
Generation Applied to Design for Remanufacture,
Proceedings of DETC’02 ASME 2002 Design Engi-
neering Technical Conferences and Computers and
In-formation in Engineering Conference, Montreal,
Que-bec, Canada.
Hill B., 1997, Innovationsquelle Natur, Technische
Universität München.
Kaiser M. K., Hashemi Farzaneh H., and Lindemann U.,
2012, An Approach to Support Searching for
Biomimetic Solutions Based on System
Characteristics and its Environmental Interactions,
International Design Conference - Design 2012,
Dubrovnik, Croatia.
Ke J., Wallace J. S., and Shu L. H., 2009, Supporting
Biomimetic Design through Categorization of Natural-
Language Keyword-Search Results, Proceedings of
the ASME 2009 International Design Engineering
Technical Conferences & Computers and Information
in Engineering Conference IDETC/CIE 2009, San
Diego, California, USA, pp. 1–10.
Ke J., Wallace J. S., Chiu I., and Shu L. H., 2010,
Supporting Biomimetic Design by Embedding
Metadata in Natural-Language Corpora, Proceedings
of the ASME 2010 International Design Engineering
Technical Conferences & Computers and Information
in Engineering Conference IDETC/CIE 2010, Montre-
BIOscrabble-ExtractionofBiologicalAnalogiesoutofLargeTextSources
19

al, Quebec, Canada, pp. 1–8.
Löffler S., 2008, Anwenden bionischer Konstruktions-
prinzipe in der Produktentwicklung, Technische Uni-
versität Carolo-Wilhelmina zu Braunschweig.
Mak T. W., and Shu L. H., 2004, Abstraction of Bio-
logical Analogies for Design, CIRP Annals - Manufac-
turing Technology, 53(1), pp. 117–120.
Nagel J. K. S., Stone R. B., and McAdams D. A., 2010,
An Engineering-to-Biology Thesaurus For
Engineering Design, Proceedings of the ASME 2010
International Design Engineering Technical
Conferences & Computers and Information in
Engineering Conference IDETC/CIE 2010, Montreal,
Quebec, Canada.
Nagel J. K. S., and Stone R. B., 2011, A Systematic
Approach to Biologically-Inspired Engineering
Design, Proceedings of the ASME 2011 International
Design Engineering Technical Conferences &
Computers and Information in Engineering
Conference IDETC/CIE 2011, Washington, DC, USA.
Pahl G., Beitz W., Feldhusen J., and Grote K. H., 2007,
Engineering Design: a Systematic Approach,
Springer-Verlag, London.
Shu L. H., 2004, Generalizing the Biomimetic Design
Process, Proceedings of CDEN/RCCI Inaugural De-
sign Conference, Montreal, Quebec, Canada.
Shu L. H., 2010, A natural-language approach to bio-
mimetic design, Artificial Intelligence for Engineering
Design, Analysis and Manufacturing, 24(04), pp. 507–
519.
Son J. (Jay), Raulf C., Cheong H., and Shu L. H., 2012,
Exploring the Collective Categorization of Biological
Information for Biomimetic Design, Proceedings of
the ASME 2012 International Design Engineering
Technical Conferences & Computers and Information
in Engineering Conference IDETC/CIE 2012,
Chicago, IL, USA, pp. 1–9.
Srinivasan V., Chakrabarti A., SAPPhIRE – An Approach
to Analysis and Synthesis, International Conference
on Engineering Design, ICED09, Stanford, CA, USA
Stone R. B., and Wood K. L., 2000, Development of a
Functional Basis for Design, Journal of Mechanical
Design, 122(December).
Stroble J. K., Stone R. B., McAdams D. A., and Watkins
S. E., 2009, An Engineering-to-Biology Thesaurus to
Promote Better Collaboration , Creativity and
Discovery, Proceedings of the 19th CIRP Design Con-
ference – Competitive Design, pp. 30–31.
Vakili V., and Shu L. H., 2001, Towards Biomimetic
Concept Generation, Proceedings of DETC’01 ASME
2001 Design Engineering Technical Conferences
Design Theory and Methodology, Pittsburgh,
Pennsylvania.
Vandevenne D., Verhaegen P.-A., Dewulf S., and Duflou
J. R., 2011, A Scalable Approach for the Integration of
Large Knowledge Repositories in the Biolog-ically-
Inspired Design Process, International Conference on
Engineering Design, ICED11, Copenhagen, Denmark.
Vandevenne D., Verhaegen P.-A., Dewulf S., and Duflou
J. R., 2012, Automated Classification into the
Biomimicry Taxonomy, International Design Confer-
ence - Design 2012, Dubrovnik, Croatia, pp. 1161–
1166.
Vandevenne D., Caicedo J., Verhaegen P.-A., Dewulf S.,
and Duflou J. R., 2012, Webcrawling for a Biological
Strategy Corpus to Support Biologically-Inspired
Design, CIRP Design 2012, A. Chakrabarti, ed.,
Springer-Verlag, London, pp. 83–92.
Vattam S. S., and Goel A. K., 2011, Foraging for In-
spiration: Understanding and Supporting the Online
In-formation Seeking Practices of Biologically
Inspired Designers, Proceedings of the ASME 2011
International Design Engineering Technical
Conferences & Computers and Information in
Engineering Conference IDETC/CIE 2011,
Washington, DC, USA, pp. 177–186.
IC3K2013-DoctoralConsortium
20
