
Automated Classification of Haematopoietic Compartments in the
Human Bone Marrow using Reservoir Computing
Philipp Kainz
1
, Harald Burgsteiner
2
, Helmut Ahammer
1
and Martin Asslaber
3
1
Institute of Biophysics, Center for Physiological Medicine, Medical University of Graz, Graz, Austria
2
Institute for eHealth, Department of Applied Computer Science,
FH JOANNEUM - University of Applied Sciences Graz, Graz, Austria
3
Institute of Pathology, Medical University of Graz, Graz, Austria
1 INTRODUCTION
Digital pathology is an emerging field in medicine
(Cross et al., 2002) and – among others – includes
sub-disciplines like telepathology, virtual microscopy
and digital image processing. As digitalization de-
vices became more precise over the past years and
whole slide imaging has been well accepted as an al-
ternative to the conventional slides, lots of new means
in the slide analysis process were revealed (Riber-
Hansen et al., 2012; Hamilton et al., 2012). Dig-
ital slides may be used in a variety of applications
like education, digital diagnostics, research, or digital
archiving (Al-Janabi et al., 2011; Riber-Hansen et al.,
2012; Hamilton et al., 2012). Every day, hundreds
of glass slides are processed manually (Huang et al.,
2011), which is a tedious and error-prone activity.
Background, Relevance and Motivation. The his-
tological examination of bone marrow is considered
in the diagnostic process of a wide range of diseases
including leukemia, anemia or lymphoma. In the cur-
rent routine diagnosis process the quantification of the
cellularity in the human bone marrow can be deter-
mined using histological sections. Though, the tech-
nology of digitized glass slides has been known for
several years, using digital images in the daily diagno-
sis process in pathology is currently being established
(Kayser et al., 2012).
The cellularity of a bone marrow specimen is ex-
pressed as percentage of the different haematopoietic
compartments. A multi-potent stem cell is the ori-
gin of all types of bone marrow cells: erythrocytes,
granulocytes, monocytes, megakaryocytes and its cor-
responding precursors as well as macrophages and
mast cells. The different bone marrow cell types can
be discriminated by morphology and spatial distri-
bution within the bone marrow (Bain et al., 2000).
Currently, histomorphometry of erythrocytopoiesis,
granulocytopoiesis, and megakaryocytopoiesis is vi-
sually performed by the pathologist. Both the over-
all quantity and an increased or decreased quantity of
haematopoietic precursors is assessed. This method
heavily relies on the experience of the pathologist.
Generally, six development stages of erythrocytes,
six of granulocytes and three of megakaryocytes are
known. A correct discrimination of the development
stages is inherently quite difficult, since the evolu-
tion is continuous. The subjective emphasis of mor-
phological criteria may lead to an under- or overesti-
mation of the cellularity of the different components
of haematopoiesis (Bain et al., 2000). Consequently,
both intra- and inter-observer variability is in cer-
tain cases significantly high, as reported from several
other research fields in digital pathology (Foss et al.,
2012; Trocchi et al., 2012; Riber-Hansen et al., 2012;
Cooper et al., 2012; Revell, 1983).
An automated detection of cell types and, as a con-
sequence thereof, the quantification of the percentage
of haematopoietic compartments would be of great-
est benefit for medical diagnostics based on histolog-
ical images. A method like this would improve ac-
curacy, sensitivity, and specificity and support the di-
agnose process quantitatively and qualitatively. Al-
though there has been a lot of progress in research,
no single quantitative method has been proposed as a
gold standard in digital pathology so far.
1.1 Computational Intelligence and
Machine Learning
Computational Intelligence (CI) refers to a sub-
branch of Artificial Intelligence (AI). Evolution-
ary algorithms, fuzzy logic, Artificial Neural Net-
works (ANN) (Haykin, 1999), Reservoir Computing
paradigms (Schrauwen et al., 2007) like Liquid State
Machines (Maass, 2010) and Echo State Networks
8
Kainz P., Burgsteiner H., Ahammer H. and Asslaber M. (2013).
Automated Classification of Haematopoietic Compartments in the Human Bone Marrow using Reservoir Computing.
In Doctoral Consortium, pages 8-18
Copyright
c
SCITEPRESS

(Jaeger, 2001), and swarm intelligence are some CI
paradigms used to facilitate “intelligent” behaviour.
Whilst these paradigms solely have been successfully
applied to real-world-problems, trends towards com-
binations of these algorithms can be observed (Ver-
straeten et al., 2007; Engelbrecht, 2007; Hassanien
et al., 2008; Yao, 1999).
CI methods can be applied to various problems
including classification, (non-linear) regression and
clustering (Bishop, 2006). Regarding the classifica-
tion the learning system tempts to develop classifica-
tion rules in order to determine a specific class out of
an input pattern. In case of regression problems in-
put data of a set A are tried to be mapped on target
values of a set B by fitting the parameters of a math-
ematical model. By doing this, future attribute values
can be predicted. Clustering is based on the idea of
letting the learning system suggest suitable classifica-
tions out of available patterns without predefining the
target class. Commonly known clustering algorithms
are (fuzzy) k-Means clustering, or Estimation Maxi-
mization (Bishop, 2006; Gonzalez and Woods, 2008).
In addition to intelligent algorithms statistical
methods, e.g. variance and correlation analyses, or
entropy are used supplementary. Those are often used
for the purpose of pre-processing raw data for cer-
tain learning algorithms. The representation of in-
put data contributes significantly to a proper perfor-
mance of intelligent algorithms. Though, poorly pro-
cessed data, e.g. inexact measurements, or noisy
data, may cause a bad learning behaviour, CI al-
gorithms are somehow error-resistant (Engelbrecht,
2007), because the “natural” variance of data can also
be learned.
The methods of CI are applied more and more to
biomedical and biochemical problem domains (Fogel,
2008; Mitra and Pal, 2005). Medical imaging is a data
intensive research area (Cooper et al., 2012; Al-Janabi
et al., 2011; Mori et al., 2008) and thus, another ad-
vantage of these methods is to be able to process
more data in less time when compared to a human.
They are suitable for automated generation, or deriva-
tion, respectively, of new knowledge out of large data
sets and take up, where human cognition is limited
in time and complexity. For instance, CI methods
are used to discover coherences in high-dimensional
data spaces (associations) or for pattern recognition
in digital images and videos (classification). Combi-
nations of different methods may yield better results
in developing a learning model or in pre-processing
the raw data (Dullin et al., 2007; Aizenberg et al.,
2001). Other CI methods for image processing have
also been proposed, like convolutional neural net-
works (Chua, 1998), pulse-coupled neural networks
(Wang et al., 2010; Kuntimad and Ranganath, 1999;
Ranganath and Kuntimad, 1999) or probabilistic neu-
ral networks (Specht, 1990), and have been success-
fully applied to image segmentation and object recog-
nition problems.
1.2 Reservoir Computing
Reservoir computing (RC) (Schrauwen et al., 2007)
refers to a quite novel paradigm in CI dealing with
separate training of recurrent neural networks (the
“reservoir”) and its readouts (Verstraeten et al., 2007;
Luko
ˇ
sevi
ˇ
cius et al., 2012). RC and its underlying the-
ory – computational neuroscience – recently became
an emerging field in information processing (Abbott,
2008; Luko
ˇ
sevi
ˇ
cius and Jaeger, 2009).
Liquid State Machines (LSMs) (Maass et al.,
2002a) are one possible implementation of the RC
paradigm and have been developed at the Graz Uni-
versity of Technology. A basic LSM architecture
(Maass et al., 2002b) consists of an input layer of
input neurons, a recurrent neural network, the neu-
ral microcircuit (NMC), of biologically realistic spik-
ing neurons (“liquid”, or “reservoir”) and a readout
layer, which can be constructed of another ANN, or
other types of neurons (Maass et al., 2002a). The
neurons within the NMC are randomly connected,
though, their initial connectivity (dynamic spiking
synapses (Maass and Markram, 2004)) is constrained
by certain probabilities, depending on whether two
neurons are of inhibitory or excitatory kind and their
spatial distance, see (Burgsteiner, 2006) for an ex-
ample model. The connections within the NMC are
not altered during the learning procedure, but just
the weights between readout layer and microcircuit
are learned within a supervised training procedure
(Goodman and Ventura, 2005). The basic princi-
ple behind LSMs is that the input layer continuously
feeds the input to the reservoir which maps the input
space non-linearly to a very high-dimensional feature
space, where – according to Cover’s theorem (Cover,
1965) – linear separability is more likely. The reser-
voir’s state is recorded over time and the readout layer
is trained on the feature space in order to accomplish a
given task with a minimal error and greatest possible
generalization capability (Bishop, 1995).
Concurrently to the LSM, Echo State Networks
(ESN) have been proposed as a similar approach, but
due to our hypothesis this project will eventually try
to implement a LSM because of a biologically more
realistic approach than other RC paradigms (ESN
(Jaeger, 2001; Jaeger et al., 2007), Backpropagation-
Decorrelation (Steil, 2004), and Temporal Recurrent
Networks (Dominey and Ramus, 2000), reviewed in
AutomatedClassificationofHaematopoieticCompartmentsintheHumanBoneMarrowusingReservoirComputing
9

(Luko
ˇ
sevi
ˇ
cius and Jaeger, 2009)). LSMs facilitate
real-time parallel computation by exploiting the non-
linear computational power of just one “liquid” and
are very well suited for temporal classification tasks
(Verstraeten et al., 2006). Since the LSM’s first oc-
currence in (Maass et al., 2002a), improvements of
the LSM architecture and training have been proposed
in (Norton and Ventura, 2006; Norton and Ventura,
2010; Hazan and Manevitz, 2010).
2 STATE OF THE ART
There has been a lot of research in AI, digital pathol-
ogy, and digital histological images (Kayser et al.,
2008a; Kayser et al., 2008b; Kayser et al., 2009;
Kayser, 2011; Molnar et al., 2003). Applications
of CI methods to images of histological specimen
yielded promising results (Sj
¨
ostr
¨
om et al., 1999).
ANN were used in combination with standard meth-
ods of image analysis for cell counting in histologi-
cal images (100× magnification, digital microscopy,
single band 8 bit grey value images). The developed
machine learning system was up to six times faster
than an experienced human, and in contrast to the
human, the system produced fewer and more con-
stant errors. The software used for cell counting was
not capable of dealing with noisy images, hence just
semi-automatic counting could be performed. Addi-
tionally, the authors found out, that the background
of an image can substantially compromise the perfor-
mance of an automated learning system. They chose
ANN for pattern recognition, because setting a sim-
ple histogram threshold and the search for grey value
peaks in an image of Hematoxylin-Eosin (HE) stained
specimen was not expedient. Some cells in the digi-
talized microscopy had multiple grey value peaks and
the threshold algorithm separated the single cell into
two distinct ones. The authors additionally tried to
train separate ANNs to determine the cell type by its
morphology, but the data was not included in this pub-
lication. Similar work has been done by (Schaberg
et al., 1992), because ANNs are able to tolerate negli-
gible image noise to a certain extent (Lin et al., 1998).
In another work (Zheng et al., 2004) directly fed
cropped image regions of 32 × 32 pixels into ANNs
of different architectures and concluded that the clas-
sification using a two-layer Feed-Forward-ANN (FF-
ANN) with shared weights yielded remarkable results
(98 − 99% correct classification rates). Others have
already experience with bone marrow material and
proposed an automated segmentation and classifica-
tion method based on features of the nuclei of white
blood cells (Shivhare and Shrivastava, 2012). Despite
their efforts and acceptable classification results using
FF-ANN they were not able to achieve automated cell
segmentation.
A huge obstacle in automated image analysis is
the pre-processing of image data for classifiers. A
common approach for reducing the dimension of a
problem domain is the representation of complex
information as features. The Principal Component
Analysis (PCA) can be used for feature extraction
(Bishop, 2006) and is applied even in other fields of
image processing for feature ranking (Zhang and Wu,
2011). Decision trees can be used for main feature ex-
traction of more abstract data like features of digital
images, too (Lu and Yang, 2009). There is no golden
rule, whether raw pixel data or more compressed in-
formation in features works best for a given problem;
both approaches yielded good results.
In histological images one can distinguish be-
tween the object space, which (probably) contains
objects (e.g. cells) and the background, containing
(probably) no objects. Frequently it is sufficient to de-
termine appropriate threshold values in the gray value
spectrum of an image in order to separate the back-
ground from the object space (Kayser et al., 2009).
One problem still remains: some objects may seem
to be connected, although they are solitary, e.g. if
they overlap in the histological section. Additional
color channels (e.g. in RGB color space) or color
space conversions are possible solutions to the prob-
lem of searching for an appropriate threshold. Com-
mon standard image processing methods like edge de-
tection algorithms, morphological operations, texture
based filters (Gonzalez and Woods, 2008) or pixel-
size filters may serve as valuable links in the pre-
processing chain for CI algorithms.
In their review about histopathological image
analysis (Gurcan et al., 2009) point out that there
is a clear need for quantitative methods in disease
grading and that computer-aided diagnosis processes
are substantial to modern processes in pathology de-
partments. Machine learning algorithms are power-
ful tools to support the daily life in digital pathology,
if properly implemented in process-oriented software.
CI has the vast potential to overcome existing barriers
and eradicate weaknesses of current standard meth-
ods. As a result, the application of its methods pro-
motes the progress in medical research.
3 RESEARCH PROBLEM
The research question addressed in this PhD project
can be separated in two domains: (i) the cell segmen-
tation and (ii) the object recognition and classifica-
IJCCI2013-DoctoralConsortium
10

tion. Classical quantitative image analysis deals with
pattern recognition based on grey level intensities in
pixels and neighbourhoods. Texture is a feature used
to partition images into regions of interest and to cate-
gorize those regions into object classes (Tuceryan and
Jain, 1998). It provides information on the spatial
arrangement of pixel colours, or intensities, respec-
tively, in an image. Both texture classification and
segmentation are common methods for pattern recog-
nition of different kinds of tissues. Texture classifi-
cation aims at matching given image regions of in-
terest with existing texture classes. Applying texture
classification solely is problematic due to the natural
variety of cell morphology. Texture segmentation fol-
lows an automated processing approach to determine
distinct texture regions within an image. Statistical
measures like the Grey Level Co-Occurrence Matrix
(GLCM), run-length statistics, contrast, entropy, vari-
ance, or energy (Gonzalez and Woods, 2008) are use-
ful, if micro-textures are the observation element of
interest. A commonly known problem when process-
ing histological images is the explicit determination
of an object (e.g. a cell) using parameters of the stan-
dard image processing, i.e. simple grey level intensi-
ties without information on their spatial arrangement
(Sj
¨
ostr
¨
om et al., 1999). Nevertheless, a final classifi-
cation of a (segmented) sample or image must always
be performed by the pathologists themselves.
Complementary to the aforementioned image pro-
cessing methods, approaches using machine learning
systems are increasingly becoming popular in image
analysis and computer vision. Several methods for
both the classical and the neural image processing ap-
proach have been proposed and applied to specific
problems in image processing domains. The auto-
mated segmentation and classification of objects in
the human bone marrow has not yet been solved satis-
factorily and digital pathology is demanding new so-
lutions for reliable decision support based on objec-
tive, robust, and reproducible results. Attempts have
been undertaken using these kind of methods in med-
ical image processing and there is progress in similar
research areas, but there is currently no solution to our
problem, or a proposed standard method, respectively.
However, there does not exist the one method
solving all problems, because each single problem re-
quires a critical view on the method to be applied due
to its characteristics (Fogel, 2008).
4 OUTLINE OF OBJECTIVES
Hypotheses. The following hypotheses are to be eval-
uated in the scope of this PhD project.
1. Computational Intelligence algorithms can be
used to solve the problem of automated relative
quantification of erythropoiesis, granulopoiesis
and megakaryopoiesis in histo(patho)logical im-
ages of stained human bone marrow specimen.
2. An appropriate biologically realistic neural net-
work can be implemented in order to classify and
quantify automatically three distinct cell types in
digital images of bone marrow.
3. New automated diagnosis methods using Com-
putational Intelligence will keep the intra- and
inter-observer variability of histological and
histopathological images at a lower and more con-
stant level when compared to humans.
Goals. The goals of the proposed PhD thesis are as
follows.
1. Evaluation of the best standard staining tech-
niques for bone marrow diagnosis and digitaliza-
tion of the histological slides.
At the beginning of the project we will focus on
chemical standard staining techniques of healthy
human bone marrow specimen. In further re-
search we also regard the examination of patho-
logical specimen. High image quality and reso-
lution is critical to succeeding image processing
and classification methods, especially in our case,
since we strive for classifying up to 13 cell classes
out of a single image.
2. Extension of the Liquid State Machine (LSM,
recurrent neural network) paradigm to the
haematopoiesis classification problem in the bone
marrow.
As described in section 1.1, the LSM has mainly
been designed for dealing with time-variant input-
and output-data. Within this project we face static,
incoherent 2D whole slide images, where experi-
enced pathologists are going to label (classify) the
cells for training. We will have to extend the LSM
paradigm in terms of finding a way to generate
temporal-like input from static data and present-
ing it to the LSM’s input layer. This stream gener-
ation process is a crucial advancement in research
and has not been proposed yet. The human vi-
sual perception and its biological signal process-
ing mechanism in the visual cortex is complex and
holds tremendous computational power. There-
fore, the information processing chain from the
retina to the brain is taken as inspiring example
for our neural network architecture.
3. Application, testing and evaluation of the adapted
Liquid State Machine on cells of the bone mar-
row.
We plan on applying the visual capability of the
AutomatedClassificationofHaematopoieticCompartmentsintheHumanBoneMarrowusingReservoirComputing
11

LSM to single images of the stained human bone
marrow and imitate the human visual perception.
The performance of the system will be evaluated
comparing the trained classifier’s quantification to
the quantification of several experienced patholo-
gists on the same image. Additionally, we will
run benchmark tests, comparing our classifier to
other, commonly used neural computation meth-
ods in image processing like Hopfield networks,
multilayer perceptrons, support vector machines
or radial basis function networks. Another goal is,
to evaluate the proposed approach to classic object
recognition methods like template matching.
5 METHODOLOGY
Major tasks of this dissertation project are:
1. the generation of the data sets for training classi-
fication algorithms, comprising
(a) digitalization of histological sections,
(b) the segmentation of the cells in the virtual
slides,
(c) data pre-processing for the algorithms, and
2. the design of the machine learning system capable
of classifying the haematopoietic compartments
within an histological image of the bone marrow.
5.1 Material and Data Acquisition
Bone marrow material is harvested from the human
iliac crest during examinations in the clinical practice
at Graz University Hospital. The material is embed-
ded in acrylate and stained at the Institute of Pathol-
ogy of the University Hospital. As a first step, and
in order to train a classifier on cell image data of the
bone marrow, we will use images of healthy bone
marrow since the intra-class cell morphology is rather
stable than in neoplastic tissue. In further experiments
we consider pathological tissue grading, too. Ex-
periments will determine suitable staining techniques.
The sections will be stained using at least the follow-
ing standard staining techniques: Hematoxylin-Eosin
(HE), May-Gr
¨
unwald Giemsa (MGG), Toluidin Blue
(TB), Gomori’s Silvering (GOM), and Periodic Acid
Schiff (PAS), where the first two are diagnostically
most conclusive for pathologists. Figure 1 shows a
cropped image of HE and MGG stained healthy hu-
man bone marrow as an example. Additional custom
staining techniques will be taken into account, if the
standard techniques are insufficient for the automated
image analysis. The glass slides are digitized at 40×
magnification using an Aperio ScanScope scanner,
20 um
Figure 1: This figure shows a scanned image of healthy
human bone marrow stained with Hematoxylin-Eosin (HE,
left), and May-Gr
¨
unwald Giemsa (MGG, right), respec-
tively, at 40× magnification.
available at the Center for Medical Research ZMF
at the Medical University of Graz. Specimen sizes
varied between 5 × 10mm and 7 × 14mm, resulting
in 24 bit, JPEG-compressed RGB images of about
25 000 × 50 000 pixels and about 200 MB per file.
5.2 Data Set Generation
Since we plan on experimenting with different super-
vised and unsupervised methods, training, validation
and test data set are required. Supervised methods
need target outputs (classes) representing the different
development stages of the haematopoietic compart-
ments. The inputs of RC approaches like ESN and
LSM are usually several temporal signals and there-
fore a couple of concurrent 1D signals. In order to
generate temporal-like 1D signals from static 2D im-
ages, we pursue the following approach.
5.2.1 Image Pre-processing and Pathway
Generation
Image pre-processing is an important step in the
preparation of raw images for intelligent methods
and is partly performed in ImageJ
1
, IQM
2
and MAT-
LAB
3
, where a large library of classical image pro-
cessing and enhancement methods is already avail-
able. As a first step, a window containing the most
significant information on the classes to be detected
is chosen from the source image and the virtual slide
1
Available from http://rsb.info.nih.gov/ij/.
2
Available from https://sf.net/projects/iqm/. This soft-
ware is developed by our research group Quantitative Mor-
phology and non-linear Methods at the Institute of Bio-
physics, Medical University of Graz.
3
Available from http://www.mathworks.com/.
IJCCI2013-DoctoralConsortium
12

is cropped to the window’s dimensions. The cells C
i
,
with i = 1, . . . , K, of interest are segmented by stan-
dard image processing methods. Hereinafter, the cell
nuclei’s centers are determined and marked (depicted
as white dots in Figure 2). These centers delineate a
plane of nodes N
i
, where the minimum Euclidean dis-
tance min({D(a, b
j
)}) between a node a and its ad-
jacent nodes b
j
determines the path to the next node.
We are going to apply certain restrictions in order to
determine a practical neighbourhood {b
j
} ⊆ {N
i
} for
node a. This successively constructs a directed graph
for subsequent extraction tasks. If it turns out that
this naive approach is insufficient, we consider using
a more sophisticated algorithm for the pathway con-
struction. Since the number of input neurons is im-
mutable after learning a special task, we have to be
flexible in dealing with the varying shape and size of
C
i
and cannot use a fixed patch size for the extraction.
Thus, the minimum circumscribing circle O
i
with ra-
dius r
i
of the corresponding C
i
is determined. Sub-
images S
i
(patches, regions of interest ROIs) of size
2(r
i
+ ξ
i
) × 2(r
i
+ ξ
i
) pixels are extracted, where ξ
i
denotes an additional object border tolerance. Each
S
i
contains only a single cell nucleus at its center. S
i
centers and O
i
centers are congruent. With three dis-
tinct cell types and its development stages we will get
up to 13 classes in total: C
i
is either a sub-class of
erythropoiesis E
c∈{1,...,6}
, granulopoiesis G
c∈{1,...,6}
,
or megakaryopoiesis M
c∈{1}
, with c as sub-class in-
dex for each cell type.
5.2.2 Class Labeling and Input Stream
Generation
The cropped virtual slides are viewed in ImageJ im-
age processing software, where experienced pathol-
ogists manually label the ROIs with the correct tar-
get class. The labeled patches are rotated centrically
by some angle α (e.g. α = 0, . . . , 359
◦
). This rota-
tion transforms the anisotropy of shape and staining
in a temporal-like input for the classifier. Due to both
varying intra- and inter-cell class shape and size, and
consequently the varying magnitude of r
i
, the patches
have to be scaled (S
i
→ S
0
i
) in order to fit the artificial
retina’s visual field
4
before rotation. Proportions of
the objects are preserved. All normalized and rotated
patches S
0
i
(α) are concatenated and form one input
stream Θ
i
of one cell. The grey value variations of all
pixels P
x,y
within the stream serve as the basis for the
required input signals for the classifier. Finally, we
get a set of input streams {Θ
i
} from a single image.
This process is repeated for each available cropped
4
The concept and architecture of the artificial retina is
explained in section 5.3.
virtual slide. The entire input stream generation pro-
cess for one cell class is depicted in Figure 2. The
complex models used in CI approaches require a huge
set of different training examples to be able to learn a
given task. Additionally, some of the examples have
to be reserved for testing and validation. The pro-
posed data set generation approach facilitates the cre-
ation of an extensive data set and enables us to cope
with intra-class variability and develop a more robust
rotation- and translation-invariant classifier. The data
set we will get out of all available virtual slides will
be divided into training, validation and test data set
(e.g. 75%, 10% and 15%).
5.3 System Architecture and Training
Procedure
Each cropped image contains lots of training exam-
ples (labelled cells) of different classes. Learning
one class from a single patch comprises (a) initializ-
ing (resetting) the reservoir, (b) feeding one encoded
stream into the input layer, (c) recording the reser-
voir’s activity in state vectors and (d) learning the
synaptic weights in the readout layer according to the
target class. Hence, we propose the following archi-
tecture.
5.3.1 Architecture
Based on the biological model of the retina, a 2D ar-
ray of n input neurons arranged in a circular field of
vision V
ret
with diameter d
ret
holds responsible for re-
ceiving the input. We will refer hereto as the artifi-
cial retina (AR). One can choose out of several neuron
models and input encodings in order to construct the
input layer of a LSM, e.g. spiking neurons or linear
neurons, and spike train generators like Poisson code
generator (Burgsteiner et al., 2007) or Bens Spiker
Algorithm (Schrauwen and Van Campenhout, 2003),
respectively. ESN on the other hand use artificial neu-
rons and do not require spike trains. The input layer is
connected to the reservoir by feed-forward synapses.
An additional input parameter, the scaling factor, is
considered to be included as a priori knowledge into
the architecture, since the patches have to be scaled in
order to fit V
ret
. The optimal reservoir architecture and
connectivity of its computational units will be deter-
mined in our experiments. The output layer is com-
posed of three to 13 readout neurons, depending on
the given task. However, it is not yet specified, which
kind of neuron is suited best for our retina model or
the output layer.
AutomatedClassificationofHaematopoieticCompartmentsintheHumanBoneMarrowusingReservoirComputing
13
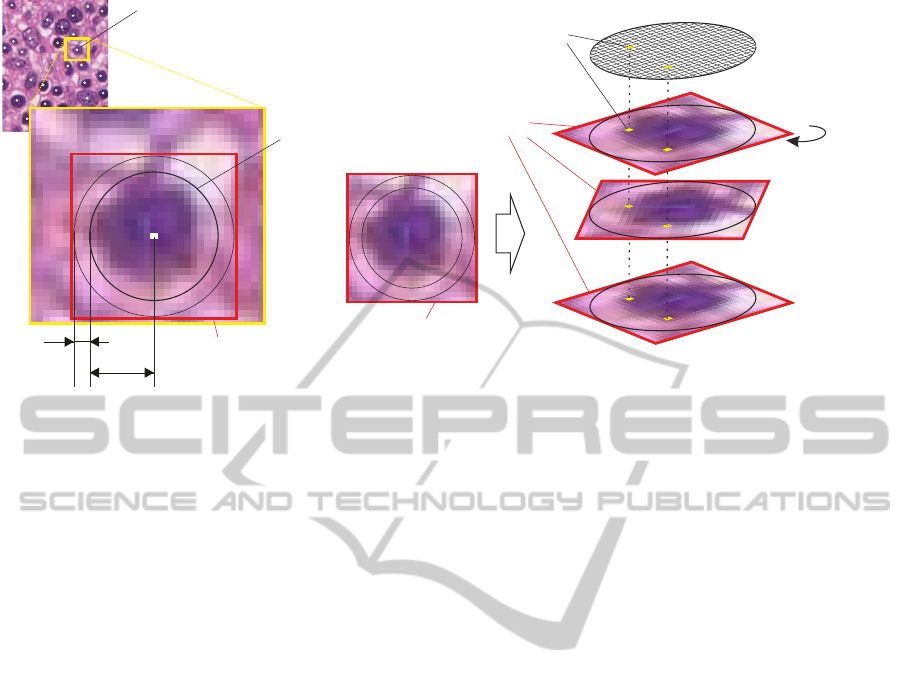
(α)
>
ξ
i
r
i
S
i
α=359°
α=45°
S´
i
V
r
et
C
i
S´
i
Θ
i
α=0°
}
O
i
P
x,y
Figure 2: This figure illustrates the generation of the image input stream Θ
i
for one cell class within a single image. This
process is repeated for each detected cell C
i
within an image.
5.3.2 Training
The initial classification task is the detection and
recognition of one of the three main cell types. A
further task is to train the classifier on all meaningful
sub-classes of each cell type.
As stated above, the cropped image is scanned
through along the pathway, continually feeding a sub-
image stream into the AR and learning the target’s
(super)class. After rotation, each image in the stream
is of different size, but since the retina is of circular
shape, all pixels outside V
ret
may be omitted. The AR
receives the input vectors u(t) (e.g. spike trains) and
forwards them to the reservoir. This causes activity
of the computational units, which is mapped to a state
vector x(t) at each sampling time-step t. From x(t),
the readout layer computes the output y(t). The ac-
tual training is done by adjusting the synaptic weights
between the reservoir and the readout neurons using
a suitable algorithm. It is not yet determined, which
training algorithm we will use eventually, but linear
regression training has already been applied succes-
sively by others (Burgsteiner et al., 2007; Hourdakis
and Trahanias, 2011) and is our first candidate. An-
other approach for multi-class discrimination is the
winner-take-all-principle, where the neuron with the
highest output activity solely determines the output
(Verstraeten et al., 2005).
5.3.3 Extensions
The cell types may express some characteristic neigh-
bourhood properties during their development, e.g.
erythroblastic islands (Bain et al., 2000). So far we
just regarded pixel grey values for training, but we
definitely consider incorporating such a priori knowl-
edge into our method. Another approach is the gener-
ation of significant image features, which will vastly
decrease the input dimensionality. Neighbourhood
conditions can be taken into account and influence –
at best enhance – the next classification output, e.g.
if both the correct classification and a distance mea-
sure to the previously correct classified cell are fed
back into the network. Using the committee paradigm
(Bishop, 1995), the training of additional classifiers
on specific cell classes will increase the accuracy of
the overall classification. With this system, we are
also able to train the algorithm not only on these three
cell types and its stages of development, but on fat and
other tissue, too. Other interesting research questions
are expected to rise as the project progresses.
5.4 Application and Evaluation
Similar to the training procedure, the starting point
for the application is a cropped histological image of
bone marrow, where the classifier is guided through
the pathway, generating classification outputs from
the observed cells. These outputs are recorded.
Thereafter, we calculate the relative amount of each
haematopoietic compartment within the image. Other
approaches like training another, subsequent (CI) al-
gorithm on the output patterns in order to decrease the
learning effort are considered as well.
The performance of the system is evaluated us-
ing several experiments and is analysed statistically.
Some experienced pathologists will independently
IJCCI2013-DoctoralConsortium
14

classify samples from the test data set and produce
the output (relative quantity of the haematopoietic
compartments) via their current standard method. In-
teresting benchmarks will be performed, namely the
comparison between the trained system and the ex-
perienced humans, in terms of overall error, intra-
and inter-observer variability, reliability, and robust-
ness. Furthermore, we will compare our classifier to
proven standard image processing methods (e.g. tem-
plate matching) as well as to other neural computing
approaches in image processing (Hopfield networks,
FF-ANN, support vector machines, radial basis func-
tions).
5.5 Software Implementation of a
Prototype Learning System
Since the image analysis software IQM, developed by
our group is written in Java, we plan to stick to this
language for our prototype system. However, MAT-
LAB is a powerful tool for signal and image process-
ing. The Neural Network Toolbox and the Reservoir
Computing Toolbox for instance are libraries provid-
ing implementations of different neural network ar-
chitectures ready to be used in some of our experi-
ments. In conjunction with other MATLAB toolboxes
we are be able to implement – at least parts of – the
prototype machine learning system. OpenCV
5
is a
large open-source library for computer vision and im-
age processing tasks. It has been extended recently
and now provides a Java binding for the C/C++ li-
brary. This advancement enables us to optimize our
pre-processing Java routines in terms of computa-
tional expenses. Furthermore, Java-ML
6
is another
machine learning library providing algorithms e.g. for
feature selection, clustering, classification, and data
filtering out-of-the-box. Java-ML has been used suc-
cessfully in several studies (Abeel et al., 2009) and is
taken into account as a candidate for particular tasks
of the prototype development. Depending on the clas-
sifier, e.g. LSM or ESN, we use other toolboxes and
simulation environments like PCSIM
7
or Oger
8
, too.
6 EXPECTED OUTCOME
In this project we are going to develop an automated
quantitative approach for the diagnosis of the cellular-
5
Available from http://opencv.org/.
6
Available from http://java-ml.sourceforge.net/.
7
Available from https://sf.net/projects/pcsim.
8
Available from http://organic.elis.ugent.be/oger.
ity of the haematopoietic compartments and its pre-
cursors in the bone marrow.
The results of our research will substantially con-
tribute to the evolution of digital pathology, namely
by providing an innovative and robust machine learn-
ing system in order to stabilize and accelerate diagno-
sis processes using state of the art information tech-
nology. Having a controlled error behaviour will de-
crease inter- and intra-observer variability and avoid
individual errors. It has to be noted that the devel-
oped machine learning system will not be restricted to
haematopoiesis classification; it can be trained on any
arbitrary task where training and test data is available.
Moreover, new insights regarding biological aspects
and knowledge on the visual perception and computa-
tion of visual stimuli may be revealed by this research.
7 STAGE OF THE RESEARCH
Data Acquisition. Essential tasks like the acquisition
and digitalization of histological sections of healthy
human bone marrow have successfully been accom-
plished. Our initial approach uses HE and MGG
stainings, but we are currently working on evaluating
other staining techniques for the sections in order to
ease the segmentation and recognition problem. Sub-
stantial time has been spent on software development
for image pre-processing. The open-source plugin
Cell Counter
9
for ImageJ has been extended to our
requirements. It is capable of recording the pathol-
ogists’ classification of the cells. So far, we have la-
beled 400 patches containing different cell types from
these images, where 55% are granulopoietic, 44% are
erythropoietic and ≈ 1% are megakaryopoietic com-
partments. More labeled patches are generated con-
tinuously and our data set is constantly growing.
System Design. First experiments are currently
planned using the ESN approach, where we manually
crop the cells from the image and omit the preceding
automated segmentation process. In order to simplify
the object recognition and discrimination, we intend
to use only two highly distinguishable classes from
our data set (early and late granulopoietic classes) in
the first place. A problem we are currently working
on is finding suitable target functions for the classes.
Approximating different constant functions are con-
sidered for the binary classification tasks. If more
and more classes are added to be learned, the func-
tions must be more discriminative. Another approach
we consider is pooled decision making in the readout
9
Available from http://rsbweb.nih.gov/ij/plugins/
cell-counter.html.
AutomatedClassificationofHaematopoieticCompartmentsintheHumanBoneMarrowusingReservoirComputing
15

layer, such that a combination of output units decides
about the categorization of the input signal.
If the results of these experiments are promising,
we plan on adapting the design to more complex prob-
lems in terms of (i) learning more than two classes
concurrently, (ii) using spiking neural networks as
reservoirs, and (iii) pursue a fully automated approach
for both segmentation and classification.
REFERENCES
Abbott, L. (2008). Theoretical neuroscience rising. Neuron,
60:490–495.
Abeel, T., Van de Peer, Y., and Saeys, Y. (2009). Java-
ML: A Machine Learning Library. Journal of Ma-
chine Learning Research, 10:931–934.
Aizenberg, I., Aizenberg, N., Hiltner, J., Moraga, C., and
Meyer zu Bexten, E. (2001). Cellular neural networks
and computational intelligence in medical image pro-
cessing. Image and Vision Computing, 19:177–183.
Al-Janabi, S., Huisman, A., and Van Diest, P. J. (2011).
Digital pathology: current status and future perspec-
tives. Histopathology.
Bain, B. J., Clark, D. M., Lampert, I. A., and Koch, S.
(2000). Knochenmarkpathologie. Blackwell, Berlin.
Bishop, C. M. (1995). Neural Networks for Pattern Recog-
nition. Oxford University Press, Inc., New York, NY,
USA.
Bishop, C. M. (2006). Pattern Recognition and Machine
Learning. Springer, New York.
Burgsteiner, H. (2006). Imitation learning with spiking neu-
ral networks and real-world devices. Engineering Ap-
plications of Artificial Intelligence, 19(7):741–752.
Burgsteiner, H., Kr
¨
oll, M., Leopold, A., and Steinbauer, G.
(2007). Movement prediction from real-world images
using a liquid state machine. Applied Intelligence,
26(2):99–109.
Chua, L. O. (1998). CNN: A paradigm for Complexity, vol-
ume 31 of World Scientific Series on Nonlinear Sci-
ence (Series A). Signapore: World Scientific Publish-
ing Company.
Cooper, L. A. D., Carter, A. B., Farris, A. B., Wang, F.,
Kong, J., Gutman, D. A., Widener, P., Pan, T. C.,
Cholleti, S. R., Sharma, A., Kurc, T. M., Brat, D. J.,
and Saltz, J. H. (2012). Digital Pathology: Data-
Intensive Frontier in Medical Imaging. Proceedings
of the IEEE, 100(4, SI):991–1003.
Cover, T. M. (1965). Geometrical and Statistical Properties
of Systems of Linear Inequalities with Applications in
Pattern Recognition. IEEE Transactions on Electronic
Computers, EC-14(3):326–334.
Cross, S. S., Dennis, T., and Start, R. D. (2002). Telepathol-
ogy: current status and future prospects in diagnostic
histopathology. Histopathology, 41(2):91–109.
Dominey, P. F. and Ramus, F. (2000). Neural network pro-
cessing of natural language: I. sensitivity to serial,
temporal and abstract structure of language in the in-
fant. Language and Cognitive Processes, 15(1):87–
127.
Dullin, C., Missbach-Guentner, J., Vogel, W. F., Grabbe, E.,
and Alves, F. (2007). Semi-automatic classification of
skeletal morphology in genetically altered mice using
flat-panel volume computed tomography. PLoS Ge-
netics, 3(7):e118.
Engelbrecht, A. P. (2007). Computational Intelligence: An
Introduction. Wiley Publishing, 2nd edition.
Fogel, G. B. (2008). Computational intelligence approaches
for pattern discovery in biological systems. Briefings
in Bioinformatics, 9(4):307–316.
Foss, F. A., Milkins, S., and McGregor, A. H. (2012).
Inter-observer variability in the histological assess-
ment of colorectal polyps detected through the NHS
bowel cancer screening programme. Histopathology,
61(1):47–52.
Gonzalez, R. C. and Woods, R. E. (2008). Digital image
processing. Prentice Hall International, Upper Saddle
River, N.J.
Goodman, E. and Ventura, D. (2005). Time invariance and
liquid state machines. In Proceedings of the Joint
Conference on Information Sciences, pages 420–423.
Gurcan, M. N., Boucheron, L. E., Can, A., Madabhushi, A.,
Rajpoot, N. M., and Yener, B. (2009). Histopatho-
logical image analysis: a review. IEEE Reviews in
Biomedical Engineering, 2:147–171.
Hamilton, P. W., Wang, Y., and Mccullough, S. J. (2012).
Virtual microscopy and digital pathology in training
and education. APMIS, 120(4):305–315.
Hassanien, A.-E., Abraham, A., Kacprzyk, J., and Peters,
J. (2008). Computational intelligence in multimedia
processing: Foundation and trends. In Hassanien, A.-
E., Abraham, A., and Kacprzyk, J., editors, Computa-
tional Intelligence in Multimedia Processing: Recent
Advances, volume 96 of Studies in Computational In-
telligence, pages 3–49. Springer, Berlin, Heidelberg.
Haykin, S. (1999). Neural Networks - A Comprehensive
Foundation. Pearson, Cambridge, London.
Hazan, H. and Manevitz, L. M. (2010). The liquid state
machine is not robust to problems in its components
but topological constraints can restore robustness. In
Proceedings of the International Conference on Fuzzy
Computation and 2nd International Conference on
Neural Computation, pages 258–264.
Hourdakis, E. and Trahanias, P. (2011). Improving the clas-
sification performance of liquid state machines based
on the separation property. In Iliadis, L. and Jayne,
C., editors, Engineering Applications of Neural Net-
works, volume 363 of IFIP Advances in Informa-
tion and Communication Technology, pages 52–62.
Springer Berlin Heidelberg.
Huang, C.-H., Veillard, A., Roux, L., Lom
´
enie, N., and
Racoceanu, D. (2011). Time-efficient sparse analy-
sis of histopathological whole slide images. Comput-
erized Medical Imaging and Graphics, 35(7–8):579–
591.
Jaeger, H. (2001). The “echo state” approach to analysing
and training recurrent neural networks - with an erra-
tum note. Technical Report GMD Report 148, Ger-
IJCCI2013-DoctoralConsortium
16

man National Research Center for Information Tech-
nology.
Jaeger, H., Luko
ˇ
sevi
ˇ
cius, M., Popovici, D., and Siewert, U.
(2007). Optimization and applications of echo state
networks with leaky-integrator neurons. Neural Net-
works, 20(3):335–352.
Kayser, K. (2011). Quantification of virtual slides: Ap-
proaches to analysis of content-based image informa-
tion. Journal of Pathology Informatics, 2:2.
Kayser, K., Borkenfeld, S., and Kayser, G. (2012). How
to introduce virtual microscopy (VM) in routine di-
agnostic pathology: constraints, ideas, and solutions.
Analytical Cellular Patholology, 35(1):3–10.
Kayser, K., G
¨
ortler, J., Bogovac, M., Bogovac, A., Gold-
mann, T., Vollmer, E., and Kayser, G. (2009). AI
(artificial intelligence) in histopathology - from image
analysis to automated diagnosis. Folia Histochemica
et Cytobiologica, 47(3):355–361.
Kayser, K., Gortler, J., Goldmann, T., Vollmer, E., Hufnagl,
P., and Kayser, G. (2008a). Image standards in tissue-
based diagnosis (diagnostic surgical pathology). Di-
agnostic Pathology, 3:17.
Kayser, K., Hoshang, S. A., Metze, K., Goldmann,
T., Vollmer, E., Radziszowski, D., Kosjerina, Z.,
Mireskandari, M., and Kayser, G. (2008b). Texture-
and object-related automated information analysis in
histological still images of various organs. Analytical
and Quantitative Cytology and Histology, 30(6):323–
335.
Kuntimad, G. and Ranganath, H. (1999). Perfect image seg-
mentation using pulse coupled neural networks. IEEE
Transactions on Neural Networks, 10(3):591–598.
Lin, W., Xiao, J., and Micheli-Tzanakou, E. (1998). A
computational intelligence system for cell classifica-
tion. In Proceedings of the 1998 IEEE international
conference on information technology applications in
biomedicine, pages 105–109.
Lu, K.-C. and Yang, D.-L. (2009). Image processing and
image mining using decision trees. Journal of Infor-
mation Science and Engineering, 25:989–1003.
Luko
ˇ
sevi
ˇ
cius, M. and Jaeger, H. (2009). Reservoir comput-
ing approaches to recurrent neural network training.
Computer Science Review, 3(3):127 – 149.
Luko
ˇ
sevi
ˇ
cius, M., Jaeger, H., and Schrauwen, B. (2012).
Reservoir computing trends. KI - K
¨
unstliche Intelli-
genz, pages 1–7.
Maass, W. (2010). Motivation, theory, and applications of
liquid state machines. In Cooper, B. and Sorbi, A.,
editors, Computability in Context: Computation and
Logic in the Real World, pages 275–296. Imperial Col-
lege Press, London.
Maass, W., Legenstein, R., and Markram, H. (2002a).
A new approach towards vision suggested by bio-
logically realistic neural microcircuit models. In
Buelthoff, H. H., Lee, S. W., Poggio, T. A., and Wall-
raven, C., editors, Biologically Motivated Computer
Vision. Proc. of the Second International Workshop,
BMCV 2002, Tuebingen, Germany, November 22–24,
2002, volume 2525 of Lecture Notes in Computer Sci-
ence, pages 282–293. Springer (Berlin).
Maass, W. and Markram, H. (2004). On the computational
power of circuits of spiking neurons. Journal of Com-
puter and System Sciences, 69:593–616.
Maass, W., Natschl
¨
ager, T., and Markram, H. (2002b).
Real-time computing without stable states: A new
framework for neural computation based on perturba-
tions. Neural Computation, 14(11):2531–2560.
Mitra, S. and Pal, S. K. (2005). Fuzzy sets in pattern recog-
nition and machine intelligence. Fuzzy Sets and Sys-
tems, 156(3):381–386.
Molnar, B., Berczi, L., Diczhazy, C., Tagscherer, A., Varga,
S. V., Szende, B., and Tulassay, Z. (2003). Dig-
ital slide and virtual microscopy based routine and
telepathology evaluation of routine gastrointestinal
biopsy specimens. Journal of Clinical Pathology,
56(6):433–438.
Mori, I., Nunobiki, O., Ozaki, T., Taniguchi, E., and
Kakudo, K. (2008). Issues for application of virtual
microscopy to cytoscreening, perspectives based on
questionnaire to japanese cytotechnologists. Diagnos-
tic Pathology, 3(Suppl 1):S15.
Norton, D. and Ventura, D. (2006). Preparing more effective
liquid state machines using Hebbian learning. In Pro-
ceedings of the IEEE International Joint Conference
on Neural Networks IJCNN’06, pages 8359–8364.
Norton, D. and Ventura, D. (2010). Improving liquid state
machines through iterative refinement of the reservoir.
Neurocomputing, 73(16-18):2893–2904.
Ranganath, H. and Kuntimad, G. (1999). Object detection
using pulse coupled neural networks. IEEE Transac-
tions on Neural Networks, 10(3):615 –620.
Revell, P. A. (1983). Histomorphometry of bone. Journal
of Clinical Pathology, 36:1323–1331.
Riber-Hansen, R., Vainer, B., and Steiniche, T. (2012). Dig-
ital image analysis: a review of reproducibility, stabil-
ity and basic requirements for optimal results. APMIS,
120(4):276–289.
Schaberg, E. S., Jordan, W. H., and Kuyatt, B. L. (1992).
Artificial intelligence in automated classification of rat
vaginal smear cells. Analytical and Quantitative Cy-
tology and Histology, 14(6):446–450.
Schrauwen, B. and Van Campenhout, J. (2003). BSA, a fast
and accurate spike train encoding scheme. In IEEE
International Joint Conference on Neural Networks
(IJCNN), pages 2825–2830. IEEE.
Schrauwen, B., Verstraeten, D., and Van Campenhout, J.
(2007). An overview of reservoir computing: theory,
applications and implementations. In Proceedings of
the 15th European Symposium on Artificial Neural
Networks, pages 471–482.
Shivhare, S. and Shrivastava, R. (2012). Automatic Bone
Marrow White Blood Cell Classfication using Mor-
phological Granulometric Feature of Nucleus. Inter-
national Journal of Scientific & Technology Research,
1(4):125–131.
Sj
¨
ostr
¨
om, P. J., Frydel, B. R., and Wahlberg, L. U. (1999).
Artificial neural network-aided image analysis system
for cell counting. Cytometry, 36(1):18–26.
Specht, D. F. (1990). Probabilistic neural networks. Neural
Networks, 3:109–118.
AutomatedClassificationofHaematopoieticCompartmentsintheHumanBoneMarrowusingReservoirComputing
17

Steil, J. J. (2004). Backpropagation-decorrelation: online
recurrent learning with O(N) complexity. In Proceed-
ings of 2004 IEEE International Joint Conference on
Neural Networks, volume 2, pages 843–848.
Trocchi, P., Ursin, G., Kuss, O., Ruschke, K., Schmidt-
Pokrzywniak, A., Holzhausen, H.-J., L
¨
oning, T.,
Thomssen, C., B
¨
ocker, W., Kluttig, A., and Stang,
A. (2012). Mammographic density and inter-observer
variability of pathologic evaluation of core biopsies
among women with mammographic abnormalities.
BMC Cancer, 12:554.
Tuceryan, M. and Jain, A. K. (1998). Texture analysis.
In Chen, C. H., Pau, L. F., and P., W. P. S., editors,
The Handbook of Pattern Recognition and Computer
Vision, chapter 2.1, pages 207–248. World Scientific
Publishing Co., 2nd edition.
Verstraeten, D., Schrauwen, B., D’Haene, M., and
Stroobandt, D. (2007). An experimental unification
of reservoir computing methods. Neural Networks,
20(3):391 – 403.
Verstraeten, D., Schrauwen, B., and Stroobandt, D. (2006).
Reservoir-based techniques for speech recognition. In
Proceedings of the World Conference on Computa-
tional Intelligence, pages 1050–1053.
Verstraeten, D., Schrauwen, B., Stroobandt, D., and
Van Campenhout, J. (2005). Isolated word recognition
with the liquid state machine: a case study. Informa-
tion Processing Letters - Special issue on applications
of spiking neural networks, 95(6):521–528.
Wang, C., Li, S., He, K., Lin, Z., and Jiang, C. (2010). Au-
tomatic image segmentation using pulse coupled neu-
ral network and independent component analysis. In
2010 International Conference on Machine Vision and
Human-Machine Interface (MVHI), pages 261 –263,
Kaifeng, China.
Yao, X. (1999). Evolving artificial neural networks. Pro-
ceedings of the IEEE, 87(9):1423–1447.
Zhang, Y. and Wu, L. (2011). Crop classification by forward
neural network with adaptive chaotic particle swarm
optimization. Sensors, 11:4721–4743.
Zheng, Q., Milthorpe, B. K., and Jones, A. S. (2004). Direct
neural network application for automated cell recog-
nition. Cytometry Part A, 57A(1):1–9.
IJCCI2013-DoctoralConsortium
18
