
Search of Possible Insertions in Bacterial Genes
Eugene Korotkov
1,2
, Yulia Suvorova
1
and Maria Korotkova
2
1
Bioinformatics Laboratory, Centre of Bioengineering Russian Academy of Sciences,
117312, prospect 60-tya Oktyabrya 7/1, Moscow, Russian Federation
2
National Nuclear Investigational University (MIFI),115522, Kashirskoe Shosse, 31,
Moscow, Russian Federation
Keywords: Triplet Periodicity, Change Points, Sequence Analysis, Genes.
Abstract: It is known that nucleotide sequences are not homogeneous and from this heterogeneity arises the task of
segmentation of a sequence into a set of homogeneous parts by the points called change points. In the work
we investigated a special case of change points in genes – paired change points (PCP). We used a well-
known property of coding sequences – triplet periodicity. The sequence that we are especially interested in
consists of three successive parts: the first and the last parts have similar triplet periodicity (TP) and the
middle part is of another TP type. We aimed to find genes with PCP and provide explanation for the
phenomenon. We developed a mathematical method for PCP detection based on new measure of similarity
between TP matrixes. Among 66936 studied genes we found 2700 genes with PCP and 6459 genes with
single change point (SCP). We suppose that PCP could be associated with double fusion or insertion events.
1 INTRODUCTION
It is widely known that nucleotide content is not
absolutely homogeneous within genetic sequences
and this heterogeneity could not be explained just by
random fluctuations (Li 1997; Elton 1974). From
this heterogeneity arises the task of segmentation of
the sequence into a set of homogeneous parts.
Analogous problem was firstly introduced in the
quality control context. It was called a “change point
problem” and a position in a sequence between two
consecutive homogeneous segments was called a
“change point” (CP) (Bhattacharya 1994). Сhange
point reflect internal changes of the process.
Many of CP finding methods were later applied
to the DNA segmentation task (Braun & Müller
1998). In this case one considers a retrospective (or
fixed) change point problem, where the entire
sequence is known prior to analysis and the task is to
find points that separate it into a set of homogeneous
and contiguous segments. The work (Braun &
Müller 1998) provides comprehensive overview and
analysis of the first change points detection methods
for DNA sequences. The first DNA segmentation
methods were based on hidden Markov models
(Churchill 1989) and walking Markov models
(Fickett et al. 1992). Later Bayesian Markov models
(Nur et al. 2009; Boys et al. 2000) and entropy
segmentation methods (Evans et al. 2010) were
introduced. A lot of methods were developed for
detecting poly-regions (regions which contain a high
occurrence of one or more nucleotides) in DNA
sequences (Papapetrou et al. 2012).
Change-points methods were used for finding
borders between coding/non-coding regions. For
instance, in the work (Bernaola-Galván et al. 2000)
entropic segmentation method based on triplet
periodicity was proposed for the task. Later the
method was improved by adding stop-codon
symbols into consideration (Nicorici & Astola
2004). This allowed authors to achieve higher
accuracy of segmentation. Similar method for
coding-region detection was developed in the work
(Deng et al. 2012) - the authors considered
dinucleotides and stop-codons.
Working with protein coding sequences we can
use their well known property, so-called “triplet
periodicity” (TP). TP is a common property of all
known living organisms and it is associated with a
gene reading frame (RF) (Frenkel & Korotkov 2008).
The feature of TP was used to distinguish coding
regions from non-coding (Shao et al. 2012).
Classification analysis of TP of genes from the
KEGG database previously showed that most of
them belonged to relatively small set of TP classes
99
Korotkov E., Suvorova Y. and Korotkova M..
Search of Possible Insertions in Bacterial Genes.
DOI: 10.5220/0004721800990108
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2014), pages 99-108
ISBN: 978-989-758-012-3
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

(about 2500 classes) and these classes may vary
greatly (Frenkel & Korotkov 2008). That led us to
the idea that if a DNA coding sequence has
fragments with different TP, this event can be
relatively easy to detect (Suvorova et al. 2012). One
can find in the sequence segments within which TP is
the same or nearly the same and between which TP
are different. And the positions between these
segments we called change points of TP. It means
that TP allows the segmentation of the gene
sequence. This work was started earlier by us
(Suvorova et al. 2012; Korotkova et al. 2011). There
are three reasons to develop a special mathematical
method for the gene segmentation task. The first one
is the relatively small size of gene sequences that
results in the small sample size statistic and forced us
to use Monte-Carlo simulations. The second is that
the triplet periodicity could change from one gene to
another (Frenkel & Korotkov 2008) as well as inside
a gene. It makes impossible to apply learning
methods such as Markov models, neural networks
and other. Third reason is related to the fact that TP is
well described by the corresponding 3×4 frequency
matrix (Frenkel & Korotkov 2008). The main subject
of the study is a gene sequence with paired change
points (PCP) of TP. This sequence consists of three
successive parts: the first and the last parts have
similar TP and the middle part is of another TP. So
one can see the first CP when going from the first
part to the second one and another CP will be found
between the second and the last part. The motivation
for this work was to improve the results of the work
(Korotkova et al. 2011) in two directions. First, we
aimed to identify pair change points without paired
reading frame shifts. The second goal was to find
PCP event with a small-size middle part (<100 b.p.).
PCP could be a marker of evolutionary sequence
formation if the sequence was formed by insertion of
one DNA sequence into another (parent) sequence or
by sequential fusions where the first and the last
fused parts have similar TP. To investigate these
sequences we introduced new measures of similarity
between TP matrixes and applied the measure of
difference between two TP matrixes that was used
before (Korotkov et al. 2003). These measures are
based on comparison of frequency matrixes of
corresponding regions. The method of PCP searching
is described in the next section. Using the method we
collected a set of genes with supposed PCP from 17
bacterial genomes. The last section presents an
analysis of the obtained results and a brief discussion.
2 METHODS AND ALGORITHMS
2.1 Data
Coding sequences for 17 genomes (Table 1) were
download from the KEGG/Genes database (Ogata et
al. 1999). These genomes together contain 69,936
gene sequences
2.2 Simulated Data
In our work we created three sets of simulated data.
The first one was dataset of homogeneous TP
sequences (denoted as Set
1
). During this simulation
we created sequences of the same length and level of
TP as in the analyzed genes. Each considered gene
sequence (S) was divided into three subsequences.
The first one (denoted as C
1
) was obtained by the
selection of symbols which were at first codon
positions in S (
( ) : 1 3 ; 0,1, 2,...( -3)/3si i n n L
).
The second sequence C
2
was generated by choosing
symbols which were at second positions
(
( ) : 2 3 ; 0,1, 2,...,( -3)/3si i n n L
), and the third
sequence C
3
was of the symbols from thirds position
(
( ) : 3 3 ; 0,1,2...( -3)/3si i n n L
). Here s(i) is the
element of sequence S. Then from sequence C
j
sequence R
j
was created by random shuffling
(j=1,2,3). And finally sequences R
j
were again
combined in one (R) in accordance to the codon
position. This simulated sequence R is of the same
length and TP level as the original gene S but after
the shuffling it became TP-homogeneous sequence.
The occurrence of PCP in the generated random
sequences could be explained only by random
fluctuations in a homogeneous sequence.
Then we simulated datasets of artificial
insertions (Set
2
) and fusions (Set
3
). We created two
simulated datasets each of 10
4
sequences. To create
these sets we randomly choose two genes from the
total dataset of 17 bacterial genomes. Then
randomly chosen parts of these genes were fused or,
in case of insertion simulation, a part of one gene
was inserted into another. These procedures were
repeated 10
4
times.
Therefore Set
1
contains sequences which TP
corresponds to the studied bacterial genes, but CP or
PCP could arise in these sequences only as a result
of random fluctuations. The volume of Set
1
was
equal to the volume of the original genes set. The
dataset Set
1
allows us to estimate the number of type
I errors (false positives) in the PCP search in genes.
There is only PCP event in each sequence from Set
2
and only SCP event in Set
3.
The Set
2
set was
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
100

constructed to estimate number of the type II errors
for the PCP search method while Set
3
allows us to
evaluate the influence of SCP events to the PCP
search in genes.
2.3 Measure of Difference between
Triplet Matrixes
We are concerned with a protein coding gene
sequence S of length L (L is divisible by three and
more than 60 b.p.). We say that there is a TP in S if
probabilities of symbols in the positions j
1
=1+3i,
j
2
=2+3i and j
3
=3+3i (i=0,1,2,…,(L-3)/3 ) differ from
the probabilities of the corresponding symbols in the
whole sequence S. In this sense TP presents in the
most of DNA sequences of length L. But only in
some sequences TP is statistically significant. It was
shown that the feature of TP is not associated with
regions with a high occurrence of one or more
nucleotides or with segmentation of genome
sequences according to GC content and gene
concentration (Melodelima et al. 2007). It is
convenient to use mutual information as a measure
of statistical significance of TP (Kullback 1997).
The mutual information (I) is computed based on TP
frequency matrix of size 4x3. The columns of the
matrix represent the positions j
1
,
j
2
and
j
3
of triplets,
and the rows represent four DNA bases. If
considered a set of random sequences S, 2I
calculated for the 4x3 matrixes would follow chi-
square distribution with six degrees of freedom
(Frenkel & Korotkov 2009). Using the chi-square
distribution one can determine a threshold value x
0
when P(2I≥x
0
)=0.05. TP of the sequence S with
2I≥x
0
one could consider as significant.
Let consider two coordinates in S: x
1
and x
2
(1≤x
1
≤x
2
≤L-l) and two corresponding regions of
length l [x
1,
x
1
+l-1) and [x
2,
x
2
+l-1). For these regions
one could calculate frequency matrixes
11 143
(,) [ (,)] MMxl mij
and
22 243
(,)[ (,)]MMxl mij
. An element of such a
matrix is a number of nucleotides of type i (i=1 for
‘a’, i=2; for ‘t’, i=3, for ‘g’ and i=4 for ‘с’), which is
in the position j of a codon (j=1,2,3), in the
considered region. For example the element m
1
(1,2)
is a number of symbols ‘t’ on the second position of
codons in the region [x
1,
x
1
+l). As a measure of
difference between two frequency matrixes we used
a value
12 112 212 312
(, ) (, ) (, ) (, )
I
MM IMM I MM IMM
(1)
where I
t
(t=1,2,3) is information measure of
difference (Kullback 1997) between the
corresponding columns of the matrixes defined as
4
12 1 1
1
( , ) (,)ln( (,))
t
i
I
MM mij mij
4
22
1
(, )ln( (, ))
i
mij mij
4
12 12
1
((,) (,))ln((,) (,))
i
mij mij mij mij
12 12
(() ())ln(() ())
s
jsj sjsj
112 2
()ln( ()) ()ln( ())
s
jsjsjsj
(2)
here
4
1
() (,)
kk
i
s
jmij
. 2I
t
has an asymptotic chi-
square distribution with three degrees of freedom
(Vinckenbosch et al. 2006). Hence
12
2( , )
I
MM has
an approximately
2
()df
and df is equal to six
because
11 2
(, )
I
MM and
212
(, )
I
MM are
independent and
312
(, )
I
MM completely determined
by
11 2
(, )
I
MM and
212
(, )
I
MM (Kullback 1997).
Then using approximation of the normal distribution
12 12
(, ) 4(, ) 2 1IM M IM M df
(3)
we obtain the value
12
(, )~(0,1)IM M N . To take
into account possible reading frame shifts after the
point x
2,
let introduce two additional matrixes for the
second region:
222 43
(,)[ (, 1)]MMxlmij
and
222 43
(,)[ (, 2)]MMxlmij
. It is useful to
note that these matrixes are the cyclic shifts of the
matrix M
2
by one or two bases correspondingly.
Using (1)-(3), one can calculate difference between
the matrix M
1
and new matrixes
2
M
and
2
M
.
Further as a measure of TP difference of two gene
regions of length l that begins at x
1
and x
2
correspondingly we used
12 12 12 12
(, ) min[( , ),( , ),( , )]Dx x I M M I M M I M M
(4)
2.4 The Similarity Measure
For similarity measure as well as in the previous
section we consider two frequency matrixes M
1
and
M
2
, which correspond to the regions of length l, and
begin at the positions x
1
and x
2
. Let us consider the
null hypothesis H
0
that the matrixes are random and
uncorrelated. Before introduce the similarity
measure between two matrixes one should
normalized them using the following element-wise
transformation
SearchofPossibleInsertionsinBacterialGenes
101

(, ) (, )
(, )
(, )(1 (, ))
kk
k
kk
mij lpij
nij
lp i j p i j
;
43
11
2
((,))((,))
(, )
kk
ij
k
mij mij
pij
l
(5)
k=1,2,
(, )~ (0,1)
k
nij N . We denoted matrixes that
obtained in the result of the transformation (5) as N
1
and N
2
. Then we constructed one more matrix
Z=[z(i,j)]
4×3
,
by multiplication of corresponding
elements of the matrixes N
1
and N
2
12
(, ) (, ) (, )zi j n i j n i j
(6)
The product of two normally distributed values
follows the distribution with density function (Craig
1936) f(z) = π
−1
K
0
(|z|) (K
0
is the modified Bessel
function of the second kind). Then for each z(i,j) one
can find probability P(z>z(i,j)) and using the inverse
function of the normal distribution calculate
corresponding value of argument of the normal
distribution y(i,j), that satisfies the condition
P(y>y(i,j))=P(z>z(i,j)). And finally we summarized
all values
43
12
11
,() (,)
ij
xSx yij
(7)
Thus, under the null hypothesis
12
(, )~ (0,6)Sx x N ,
where N(0,6) is the normal distribution with and the
value
12
((0,6) (, ))PN Sx x shows the probability
of randomness of the matrixes similarity. We tested
the distribution of
12
(, )~ (0,6)Sx x N using random
matrixes. If
12
(, )Sx x is sufficiently large, then the
probability, that similarity of two matrixes is
random, becomes low and the hypothesis about
random similarity of matrixes should be rejected.
2.5 Method for PCP Detection
Let introduce a set of points in S:
(1)1
k
xstepk; k = 1,2…K. For each position
x
k
we calculated matrixes M(x
k
,l). Totally
()/ 1KLlstep
matrixes were calculated in S
(the length of considered regions was defined as
l=60 and the step size as step=9). Then K matrixes
were compared with each other and two big matrixes
Sim=[sim(i,j)]
K×K
and Dif=[dif(i,j)]
K×K
were
constructed as:
(, ) ( , )
(, ) ( , )
ij
ij
s
im i j S x x
dif i j D x x
(8)
The elements of the matrix Sim that were
calculated using equation (7) reflect similarity and
the elements of Dif, that were calculated using (4)
reflect difference between corresponding regions.
Then for arbitrary values k
1
and k
2
(
12
1 kk K)
we calculated
12
11 2 12
121 2 122
22
11 2
11
11
(, ) (,)
(, ) (, )
(, ) (, )
(, )
ik jk
ikk jk ikk jK
kikk jk kikk jK
kjKkjK
Wkk simij
rdifij simij
s
im i j r dif i j
sim i j
(9)
To illustrate the idea of the equation (9) let assume
that the sequence S has an insertion of different TP of
length multiple to three between the positions
corresponding to k
1
and k
2
(the case of insertion not
divisible by three is described further). In this case
the first, fourth and sixth terms of equation (9) reflect
the similarity of the triplet periodicity within the
intervals (1,k
1
), (k
2
,k
1
) and (k
2
,K), respectively. The
second and fifth terms of equation (9) reflect the
difference between the TP of the intervals (1, k
1
) and
(k
2
, k
1
), and (k
2
, k
1
) and (k
2
, K), respectively. The third
term of equation (9) reflects the similarity of the TP of
the intervals (1,k
1
) and (k
2
,K). The coefficient r was
found to balance the contributions of difference and
similarity measures in the final value. On the test set
of artificial sequences with PCP (Set
2
) the r value was
chosen to maximize PCP finding (r=7). To take into
account an overall homogeneity of the considered
sequence we used the following correction
2
11
(, )
iK jK
Wsimij
(10)
Equation (10) reflects a case of homogeneous
sequence without insertions. Given this correction
for PCP search the next equation was used:
12 112 2
(, ) (, )Wk k W k k W
(11)
The calculations of W were performed for all
possible combinations of k
1
and k
2
in S. And the
positions where W reached its maximum
12
max 1 2
,
max( ( , ))
kk
WWkk
were found. Then we need to
define whether this maximum value is significant.
2.6 Determine Statistical Significance
To determine the statistical significance of W
max
for
every considered gene we simulated 500
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
102

homogeneous sequences (see materials, Set
1
). For
each simulated sequences the corresponding value of
W
max
was determined (see previous section). From
the simulated set mean
max
W and standard variance
max
()W
were calculated and finally for S we found
the statistic
max
max
max
()
WW
Z
W
(12)
In our analysis we also considered possible reading
frame shift after the second change point (this is a
case of inserts of length that is not divisible by
three). In order to consider a case of shift by one or
two positions one should use left region frequency
matrix corresponding to the second or third reading
frame instead of first in the third term of equation
(9). Let denote Z value which is corresponds to the
non-reading shift case as Z
1
, case of one-position
shift as Z
2
, and in case of shift by two positions as
Z
3
.
Because of triplet structure of real genes Z value
(equation (12)) does not follow normal distribution,
so the thresholds for Z
1
, Z
2
и Z
3
have to be found
empirically using additional simulations.
2.7 Search of Single Change Points
(SCP) of Triplet Periodicity
It is important to note that gene sequences with SCP
(Suvorova et al. 2012) could give values Z
1
, Z
2
, or Z
3
greater than the corresponding thresholds. Therefore,
each gene where PCP was found should be
additionally tested for SCP presence before the final
conclusion. Searching process of the SCP is similar
to the process of PCP search that described in the
section 2.3 but here only one coordinate k
1
is
considered and the value
11 2
(, )Wkk , is defined as
11
()Wk :
11
11 1 1
11
11
1
() (,)
(, ) (, )
ik jk
ikk jK k iKk jK
Wk simij
rdifij simij
(13)
Then for equations (10-12) were used and instead of
W(k
1
,k
2
) was used W
1
(k
1
) in formula (11). For SCP
value Z was redesignated as V in formula (10).
2.8 Determine threshold Values
To determine statistical significance of found PCP
and SCP we examined the dependencies of 1-F
Z
(z)
from Z
1
, Z
2
, Z
3
for PCP and 1-F
V
(v) for SCP cases.
Here F
Z
(z) is the distribution functions for Z
1
, Z
2
,
and Z
3
and
F
V
(v) is the distribution function for V.
To build these distribution functions we created 100
independent Set
1
sets (each real sequence was
shuffled 100 times according to procedure described
in the Section 2.2.). Then the distribution functions
were calculated for mean values of Z
1
, Z
2
, Z
3
and V.
We chosen one threshold value Z
0
for PCP and SCP
events so that the maximum of
1
Z0
1- ( )
F
Z ,
2
Z0
1- ( )
F
Z ,
3
Z0
1- ( )
F
Z and
0
1- ( )
V
F
Z constituted no
more than 18%. The value of Z
0
was equal to 3.8.
So the cases where V was the maximum
(
0
,1,2,3
i
VZ Zi ), were considered as SCP
events. And only the genes where one of Z
i
was
higher than V
0
()
i
Z
VZ , were considered as
containing PCP.
2.9 Contour Plots of TP Difference
in Genes
To illustrate TP distribution of different part of a
gene sequence we used contour plots of measure of
difference
12
(, )Dx x (equation (3)) between regions
of S of length l. Varying independently coordinates
x
1
and x
2
along the sequence (x
1
=1+3i, x
2
=1+3j,
i=0,1,2,…,(L-3)/3, j=0,1,2,…,(L-3)/3, i and j are
changed independently of each other), we calculated
matrixes M
1
and M
2.
Then
we calculated
12
(, )
I
MM
according to the formula (1) and
12
(, )
I
MM
according to the formula (2). Then the contour plot
was built to represent the dependence of
12
(, )
I
MM
on x
1
and x
2
. Such contour plots are symmetric about
the main diagonal. The darker color of a certain
region on the plot, the greater difference of the
corresponding region’s TP from TP of the rest of the
sequence. So one can see the region between two CP
that has another TP than the surrounding regions
have.
2.10 BLAST Analysis
To investigate the possible causes of PCP we
denoted the scheme of a sequence S with PCP as
S(L)=S
1
+S
2
+S
3
11
[0, ]SSCP
;
212
[, ];SCPCP
32
[,]SCPL
where CP
1
and CP
2
are the
coordinates of the first and the second CP in gene.
Now we may consider two possible ways of
evolution formation of the sequence S. The first one
is that S was formed by the insertion of sequence S
2
into a parent sequence (S
1
+S
3
). And the second
SearchofPossibleInsertionsinBacterialGenes
103
hypothesis is that S was created by two sequential
fusion events and the subsequences S
1
, S
2
and S
3
initially belonged to three different sequences (but S
1
and S
3
had similar TP).
To test both hypotheses we performed a search
of potential ancestral sequences of the sequence S by
special similarity search. Under the first hypothesis
we looked for the sequence that is similar to S
1
and
S
3
but has no central part S
2.
Under the second
hypothesis one can discover genes which are similar
to only one of the region S
1
, S
2
or S
3
. But to consider
a significant result we required the existence of at
least two regions with proper similarity (S
1
, and S
2
or
S
2
and S
3
or S
1
and S
3
) (or, in the best case, found all
three) in different sequences.
We used BLAST (Altschul et al. 1990) (option
blastx) with the E-value cutoff 0.001. BLAST
scanning was performed on a set of proteins from
the Swiss-Prot database (Boeckmann et al. 2003)
(531473 protein sequences). For each query
sequence we looked for alignments corresponding to
one of the hypotheses. Of course, CP coordinate
defined by our method could not be considered as
exact so we introduced an error interval in
comparing the coordinate of obtained alignment and
CP coordinate. The error was equal to 5% of the
length of a query sequence.
2 RESULTS
3.1 Simulated Dataset
Firstly we made the control search of the PCP in
artificial periodic sequence. We took the periodic
sequence (atg)
160
and analyzed it by developed
algorithms. In this case it is impossible to identify
any CP. Then we took the sequence of the gene of
the chitosanase from B.subtilis genome
(KEGG:BSU26890). In this gene PCP was not found
too. Then we made an insertion of fragment of 180
nucleotides with another triplet periodicity after
240
th
position of the gene. The contour plot of new
generated sequence is shown in Fig.2. One can see
the great difference between TP in the interval from
240 to 400 nt and triplet periodicity of others parts
of the sequence.
We used Set
1
and Set
2
sets to determine levels of
type I and II errors. In the first case there were 486
sequences with PCP (with level 3σ = 72). This level
was found by the analysis of 100 different Set
1
datasets. The number of type I errors for the stated
threshold (Z
0
= 3.8) constituted about 18% from the
total number of PCP found in real bacterial genes
Figure 2: Contour plot of difference of TP in gene coding of
the chitosanase from B.subtilis genome (KEGG:BSU26890)
with artificial insertion of 180nt. length after 240
th
nt. One
can see that the region from ~200 b.p. to ~300 b.p. has
different TP matrix than another sequence.
(see 3.2). The level of 18% was selected to compare
the number of PCP with the results of our previous
works (the number of insertions that was estimated
by the other method (Korotkova et al. 2011) and the
number of reading frame shifts (Korotkov &
Korotkova 2010).
We used the Set
2
set to evaluate type II error rate.
The results were the following: totally 8018 cases
were determined by the program. In 6306 cases the
results were post defined as meaning SCP and
remain 1712 cases were meaning PCP. Since the
total size of Set
2
was 10
4
sequences then the level of
type II error constitutes about 83%. The results of
the study of Set
2
demonstrate, that the method
determined only the lowest border of the possible
PCP number (because of 83% type II error rate). The
test also demonstrates that considerable part of SCP
cases found in the work could be actually PCP
events.
Last, we estimated the contribution of SCP into a
PCP number. In the Set
3
dataset 7566 cases were
defined as SCP and only 127 as PCP. This means
that about 1,3% of SCPs would be found by the
program as PCPs. Our previous results (Suvorova et
al. 2012) showed that about 10% of genes contain
SCP, which in present case can be estimated as
7x10
3
genes. The number of SCP defined as PCP by
the method should be about 90 cases from the total
set. This means that we were quite accurate in PCP
detection among the real SCP cases.
3.2 Real Dataset
Then we analyzed genes from our main set from 17
bacterial genomes (see Table 1
for details). Totally we
found 9159 gene sequences where one of Z
i
was greater
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
104

than the corresponding threshold. Subsequent analysis
revealed in this set 6459 genes with SCP (about 10% of all
studied genes, this value is consistent with our earlier
results obtained in work (Suvorova et al. 2012) and 2700
genes with PCP.
Table 1: Total Change-Points Statistic
Genome SCP PCP,
shift 0
PCP,
shift 1
PCP,
shift 2
A.butzleri 227 50 23 20
A.vinelandii_Ent 477 92 71 64
B.avium 232 72 32 14
B.mallei 847 150 94 87
B.subtilis 444 114 49 20
E.coli 357 70 35 32
L.fermentum 170 41 15 19
M.capsulatus 281 78 25 26
P.aeruginosa 635 142 96 98
S.aureus_COL 221 51 17 18
S.enterica_Choler
aesuis
417 88 60 33
S.pneumoniae 150 29 13 8
S.sonnei 396 71 35 30
S.typhimurium 392 95 50 43
V.cholerae 246 48 31 17
X.campestris 604 91 63 33
Y.pseudotuberculo
sis_YPIII
363 80 48 19
The list of used bacterial genomes with corresponding numbers of
found paired and SCP.
Genes with the SCP were described in detail in our
work (Suvorova et al. 2012) and in there we found
that SCP could be associated with fusion event. So
here we just compared corresponding genomes
results. In the previous work 5843 genes (at level of
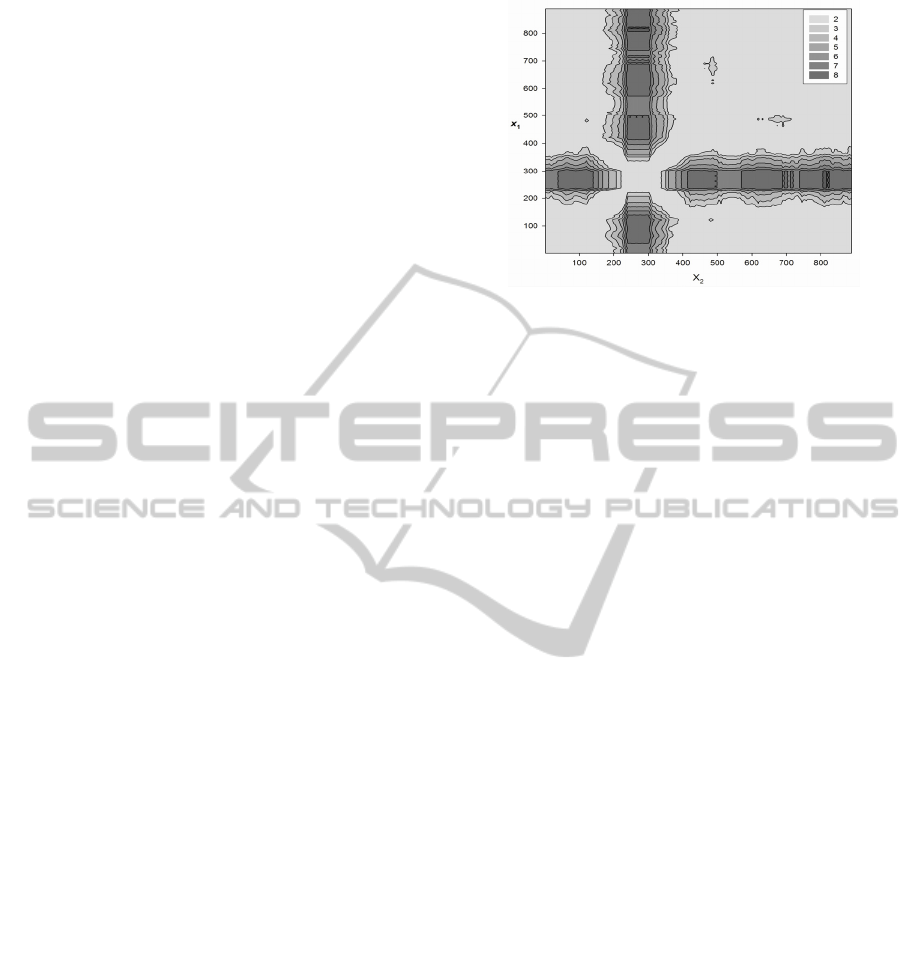
Figure 3: Contour plot of difference of TP in gene
sequence that coding the glycerol-3-phosphate permease
in B.subtilis genome (KEGG:BSU02140). One can see the
paired change points in the positions ~600 and ~700 nt.
18% false positives) with SCP were found in the
same 17 genomes and ~ 50% are in the same genes
as in the current work. The difference can be
explained by the fact that early to identify the CP we
took into account only the difference between the TP
matrixes. In some cases it could lead to detection of
shifts of the TP phase as the CP (Suvorova et al.
2012). In this study we additionally used new
similarity measure and it allowed us to obtain more
accurate results in the SCP search.
The main interest to us in this work was in the
set of 2700 genes with PCP. This number constitutes
about 4% of the sample size. Example of the gene
with PCP are shown in Fig.3. This figure
demonstrate the sequences with TP fragment
different from all other parts of the genes.
3.3 Blast Results
We performed BLAST analysis for each from 2700
sequence with PCP searching for double fusion or
insertion hypothesis corresponding alignments. For 73
sequences we found proper alignment corresponding
to the insertion hypothesis.
3.4 Insertion Database
We compared the results with online database of
insertion in protein structures (Aroul-Selvam et al.
2004). We downloaded 2137 PDB that contain
insertions and for 1676 of them corresponding DNA
sequences were found using online server
(Hovmoller & Zhou 2004). After removing the
redundancy there were 232 sequences. We tested
DNA sequences from this non-redundant set for CP
presence using our program. Significant CP cases
were found in 55 cases (35 cases of SCP, 9
sequences contain PCP without shift, 6 PCP with
shift equal one and in 5 cases PCP with shift on two
bases). Found results (20 from 232 cases) are
comparable to those obtained on simulated data
(1712 from 10
4
). The difference in the results is
effect of different distribution of TP classes in
simulated and real datasets.
4 DISCUSSION
AND CONCLUSIONS
We developed the method for finding paired and
single change points in coding sequences and the
program for visualisation of such events. The
analysis using the method was performed on both
simulated and real datasets of 17 bacterial genomes.
SearchofPossibleInsertionsinBacterialGenes
105

Our study demonstrated that about 10% of
investigated coding sequences contain SCP and
about 4% - PCP. The number of SCP is comparable
with the results of our previous work (Suvorova et
al. 2012). The results on simulated
fusions/insertions sets showed that the number of
SCP falsely detected as PCP should not be greater
than 2% of found cases. In the same time rely on the
simulation results we can conclude that the method
determined only the lowest border of the possible
PCP number (number of false negative error ~83%)
because most of them could be falsely detected as
SCP.
We found alignment-based confirmation of
relation between PCP and fusion/insertion events
only for the minor part of genes with PCP (~13%)
(see Section 3.3.). In our opinion, there are several
reasons to explain this difference. The first one is
that used database is not a comprehensive collection
of existing and existed amino acid sequences. So the
parental sequences (that were involved into
fusion/insertion events) could be absent in the
Swiss-Prot database. That’s why some of these
events could be missed by the alignment-based
search. Secondly during long evolutionary period
after fusion or insertion event occurred sequences
could be lost from present day genomes or greatly
changed so the programs could not detect similarity.
So the alignment-based methods could detect only a
small part of actually produced PCP. So, our method
could provide an additional approach for prediction
of such events.
Performed BLAST search with the same
parameters on Trembl database we obtained slightly
different results. For 86 sequences we found proper
alignment corresponding to the insertion hypothesis.
In case of double fusion hypothesis: for 34
sequences with PCP all three supposed ancestral
genes were found (similar to S
1
, S
2
and S
3
) and for
301 sequences alignments for two of three parts
were found. Despite the high level of the type II
error, the method is seems to be more effective than
alignment-based methods for detection of insertions
and paired fusions.
The modeling process lets us to conclude that
found change points in genes could not be the result
of random fluctuations. Besides the change points
could not be the result of changes in protein
structure (if it was true CP would be found
practically in every gene sequence). We suppose that
the change points are the reflection of evolution
events like fusions and insertions. The additional
BLAST testing showed that found cases of PCP
could reflect double fusion and insertion events (but
using the results we could not estimate the
quantitative contribution of these processes into PCP
formation).
It is interesting to note that we found PCP cases
with a reading frame shift. Most likely, this
phenomenon could be explained by the insertion of
DNA fragments of length not multiple of three
bases. In this work we found less than half of
insertions of DNA fragment with a length not a
multiple of three that we found before in work
(Korotkova et al. 2011). This could be due to the
fact that in the present study we considered the PCP
events and in previous work we searched for pair
phase shifts of TP. These pair TP phase shifts can
occur without any insertions but by the way of
double shifts of the reading frame. Thus, the
previous results took into account both insertions of
DNA fragments and the pair shifts the reading
frame. It implies that only about 1350 genes from
the total 2809 genes with insrtion found in our
previous work (Korotkova et al. 2011) contained
PCP. So the rest (2809-1350=1449) genes contained
paired phase shifts of TP (2,1% of the total analyzed
set). This result seems to be realistic since it lower
than the number of genes with a single TP phase
shift (3.6%) found in the work (Korotkov &
Korotkova 2010). Actual number of single phase
shifts of TP may be less than 3.6% since some cases
of paired phase shift of TP could be detected in
(Korotkov & Korotkova 2010) as a single. This
result does not related to the lack of mathematical
method developed in the work (Korotkov &
Korotkova 2010), but rather shows that the statistical
significance of pair phase shifts may be above the
threshold, but separately threshold can overcome
only one phase shift.
Also in contrast to the method proposed in the
work (Korotkova et al. 2011) our new method could
detect short insertions (<100 bp). In the previous
work this short regions were merged and considered
as SCP. That’s why we found additionally 1350
cases of PCP. This is not a surprising result because
short insertion may have less impact on the protein
structure and therefore higher chances to remain in
the gene.
A mathematical method based on Jensen–
Shannon divergence proposed in works (Bernaola-
Galván et al. 2000; Li et al. 2002) is most similar to
our approach. The method is devoted to distinguish
coding sequences from non-coding based on
presence/absence of TP in a region. Authors
introduced 12-dimentional vector (Li et al. 2002)
which is an equivalent to our TP matrix. But the
Jensen–Shannon divergence that was used to
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
106

compare the vectors computed for the subsequence
to the left and the subsequence to the right of the
pointer could not detect the difference between two
TP matrixes. Therefore in the work we introduced a
new mathematical method to detect PCP based on
measures of similarity and difference between TP
matrixes.
The method could reveal the fusion and
insertions events in genes without any additional
information. Study of sequences with artificial
insertions/fusions and distribution of TP among
genes inside genome support the idea that not all
cases of insertions or fusions could be found using
the TP changes. Only fusions/insertions of
sequences with different TP matrixes would lead to
TP change points. We suppose that real number of
genes formed by insertions or fusions events could
be 5-7 greater than we obtained in the work. Now it
is difficult to say whether the function of the protein
was changed after these events and whether such
events led to creation of new genes and new
biological functions of the encoded proteins. Some
answers to the question could be found after the
experimental work.
REFERENCES
Altschul, S. F. et al., 1990. Basic local alignment search
tool. Journal of molecular biology, 215(3), pp.403–410.
Aroul-Selvam, R., Hubbard, T. & Sasidharan, R., 2004.
Domain insertions in protein structures. Journal of
molecular biology, 338(4), pp.633–641.
Bernaola-Galván, P. et al., 2000. Finding borders between
coding and noncoding DNA regions by an entropic
segmentation method. Physical Review Letters, 85(6),
pp.1342–1345.
Bhattacharya, P., 1994. Some aspects of change-point
analysis. In Carlstein, E., Müller, H.-G., Siegmund, D.
(eds.), Change Point Problems, IMS Lecture Notes -
Monograph Series, 23(1980), pp.28–56.
Boeckmann, B. et al., 2003. The SWISS-PROT protein
knowledgebase and its supplement TrEMBL in 2003.
Nucleic acids research, 31(1), pp.365–370.
Boys, R. J., Henderson, D. A. & Wilkinson, D. J., 2000.
Detecting homogeneous segments in DNA sequences
by using hidden Markov models. Journal of the Royal
Statistical Society: Series C (Applied Statistics), 49(2),
pp.269–285.
Braun, J. V & Müller, H.-G., 1998. Statistical methods for
DNA sequence segmentation. Statistical Science, 13(2),
pp.142–162.
Churchill, G. A., 1989. Stochastic models for heterogeneous
DNA sequences. Bulletin of mathematical biology,
51(1), pp.79–94.
Craig, C. C., 1936. On the frequency function of xy. he
Annals of Mathematical Statistics, 7(1), pp.1–15.
Deng, S. et al., 2012. Detecting the borders between coding
and non-coding DNA regions in prokaryotes based on
recursive segmentation and nucleotide doublets
statistics. BMC Genomics, 13(Suppl 8), p.S19.
Elton, R. A., 1974. Theoretical models for heterogeneity for
base composition in DNA. Journal of Theoretical
Biology, 45(2), pp.533–553.
Evans, G. E. et al., 2010. Estimating Change-Points in
Biological Sequences via the Cross-Entropy Method.
Annals of Operations Research, 189(1), pp.155–165.
Fickett, J. W., Torney, D. C. & Wolf, D. R., 1992. Base
compositional structure of genomes. Genomics, 13(4),
pp.1056–1064.
Frenkel, F. E. & Korotkov, E. V, 2008. Classification
analysis of triplet periodicity in protein-coding regions
of genes. Gene, 421(1-2), pp.52–60.
Frenkel, F. E. & Korotkov, E. V, 2009. Using triplet
periodicity of nucleotide sequences for finding potential
reading frame shifts in genes. DNA research: an
international journal for rapid publication of reports on
genes and genomes, 16(2), pp.105–14.
Hovmoller, S. & Zhou, T., 2004. Protein shape strings and
DNA sequences.
Korotkov, E. V et al., 2003. The informational concept of
searching for periodicity in symbol sequences.
Molekuliarnaia Biologiia, 37(3), pp.436–451.
Korotkov, E. V & Korotkova, M.A., 2010. Study of the
triplet periodicity phase shifts in genes. Journal of
integrative bioinformatics, 7(3).
Korotkova, M. A., Kudryashov, N. A. & Korotkov, E. V,
2011. An approach for searching insertions in bacterial
genes leading to the phase shift of triplet periodicity.
Genomics, proteomics & bioinformatics, 9(4-5),
pp.158–70.
Kullback, S., 1997. Information Theory and Statistics. S.
Kullback, ed., New York: Dover publications.
Li, W. et al., 2002. Applications of recursive segmentation
to the analysis of DNA sequences. Computers &
chemistry, 26(5), pp.491–510.
Li, W., 1997. The study of correlation structures of DNA
sequences: a critical review. Computers chemistry,
21(4), pp.257–271.
Melodelima, C., Gautier, C. & Piau, D., 2007. A markovian
approach for the prediction of mouse isochores. Journal
of Mathematical Biology, 55(3), pp.353–364.
Nicorici, D. & Astola, J., 2004. Segmentation of DNA into
Coding and Noncoding Regions Based on Recursive
Entropic Segmentation and Stop-Codon Statistics.
EURASIP Journal on Advances in Signal Processing,
2004(1), pp.81–91.
Nur, D. et al., 2009. Bayesian hidden Markov model for
DNA sequence segmentation: A prior sensitivity
analysis. Computational Statistics & Data Analysis,
53(5), pp.1873–1882.
Ogata, H. et al., 1999. KEGG: Kyoto Encyclopedia of
Genes and Genomes. Nucleic Acids Research, 27(1),
pp.29–34.
Papapetrou, P., Benson, G. & Kollios, G., 2012. Mining
poly-regions in DNA. International journal of data
mining and bioinformatics, 6(4), pp.406–28.
SearchofPossibleInsertionsinBacterialGenes
107

Shao, J., Yan, X. & Shao, S., 2012. SNR of DNA sequences
mapped by general affine transformations of the
indicator sequences. Journal of Mathematical Biology.
Suvorova, Y.M., Rudenko, V.M. & Korotkov, E. V, 2012.
Detection change points of triplet periodicity of gene.
Gene, 491(1), pp.58–64.
Vinckenbosch, N., Dupanloup, I. & Kaessmann, H., 2006.
Evolutionary fate of retroposed gene copies in the
human genome. Proceedings of the National Academy
of Sciences of the United States of America, 103(9),
pp.3220–3225.
BIOINFORMATICS2014-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
108
