
Detection of Prostate Abnormality within the Peripheral Zone using
Local Peak Information
Andrik Rampun
1
, Paul Malcolm
2
and Reyer Zwiggelaar
1
1
Department of Computer Science, Aberystwyth University, Aberystwyth SY23 3DB, U.K.
2
Department of Radiology, Norfolk & Norwich University Hospital, Norwich NR4 7UY, U.K.
Keywords:
Computer Aided Detection of Prostate Cancer, Peak Detection, Prostate Abnormality Detection, Prostate MRI.
Abstract:
In this paper, a fully automatic method is proposed for the detection of prostate cancer within the peripheral
zone. The method starts by filtering noise in the original image followed by feature extraction and smoothing
which is based on the Discrete Cosine Transform. Next, we identify the peripheral zone area using a quadratic
equation and divide it into left and right regions. Subsequently, peak detection is performed on both regions.
Finally, we calculate the percentage similarity and Ochiai coefficients to decide whether abnormality occurs.
The initial evaluation of the proposed method is based on 90 prostate MRI images from 25 patients and 82.2%
(sensitivity/specificity: 0.81/0.84) of the slices were classified correctly with 8.9% false negative and false
positive results.
1 INTRODUCTION
In 2013, approximately 239,000 American men were
diagnosed with prostate cancer (more than 20,000 in-
creased compared to 217,730 cases in 2010 (Tempany
and Franco, 2012)). In the United Kingdom, over
40,000 cases are reported annually with more than
10,000 deaths (PCUK, 2013). Generally, there are
several well known clinical diagnostic tests such as
prostate-specific-antigen (PSA) level (Brawer, 1991),
digital rectal examination (DRE) (Shirley and Brew-
ster, 2011), transrectal ultrasound (TRUS) (Aus et al.,
2008) and biopsy tests (Roehl et al., 2002). Nevethe-
less, prostate cancer too often goes undetected as the
sensitivity and specificity of these techniques could be
improved and could have complications (Tidy, 2013;
Kenny, 2012; Choi et al., 2007; Tempany and Franco,
2012). Prostate magnetic resonance imaging (MRI)
can provide non-invasive imaging and in combina-
tion with computer technology can provide a detec-
tion tool which has the potential to improve the ac-
curacy of clinical diagnostic tests (Ampeliotis et al.,
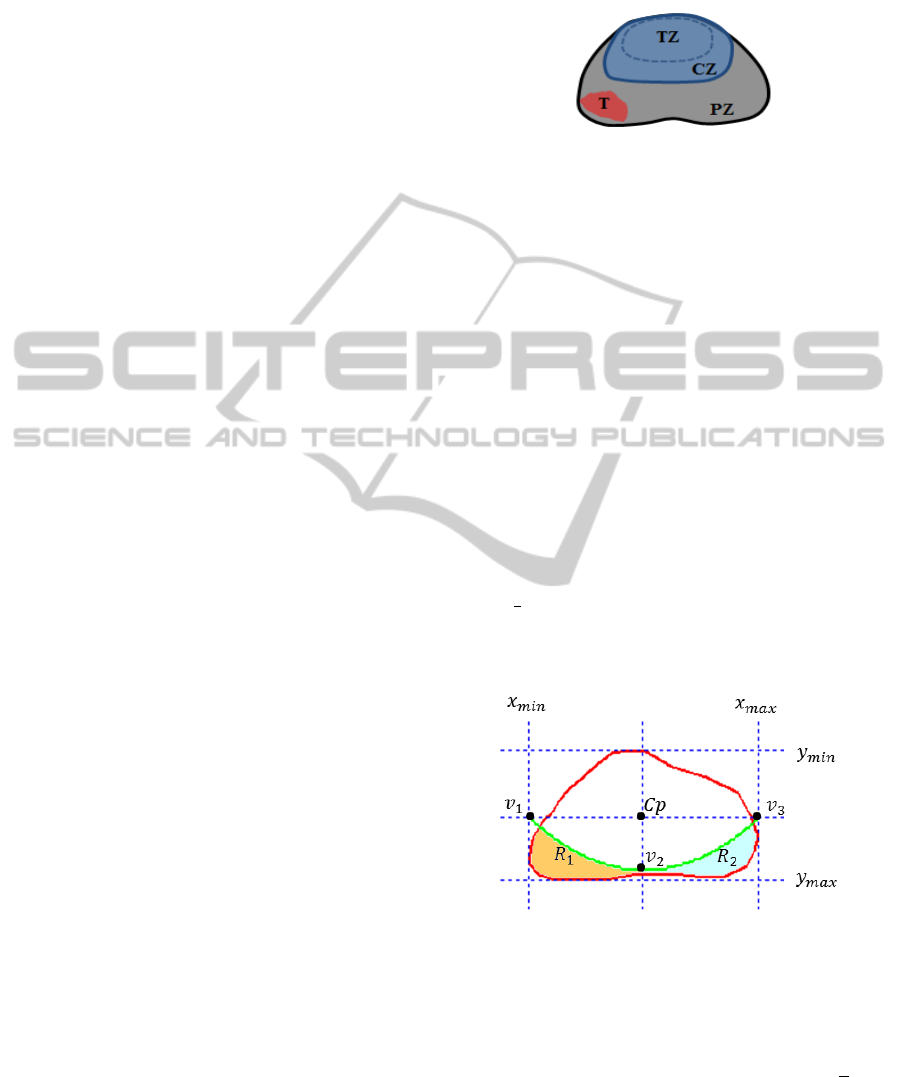
2007). Figure 1 shows an example prostate MRI im-
age with its ground truth delineated by an expert radi-
ologist.
This research aims to develop a computer aided
diagnosis (CAD) tool for prostate cancer by compar-
ing data peaks between the left and right regions of
the peripheral zone (PZ). Peak detection methods are
Figure 1: The ground truth of prostate gland, central zone
and tumor are represented in red, yellow and green, respec-
tively.
popular in many signal processing applications. How-
ever, not much work has been done applying such
technique to the detection of prostate cancer based
on MRI analysis. Limited number of methods in
the literature attempted to use peak values in detect-
ing prostate abnormality such as (Vos et al., 2010),
(Reinsberg et al., 2007) and (Choi et al., 2007). A
method proposed by (Vos et al., 2010) uses a peak
detector to select abnormal regions from the obtained
likelihood map constructed during the voxel classifi-
cation stage. Subsequently, they perform automatic
normalisation and histogram analysis for each of the
abnormal regions before calculating malignancy us-
ing a supervised classifier. On the other hand, (Reins-
berg et al., 2007) uses peaks information together
with a apparent diffusion coefficient map to detect
abnormality within the prostate. Another study by
(Choi et al., 2007) shows that peak infromation has
510
Rampun A., Malcolm P. and Zwiggelaar R..
Detection of Prostate Abnormality within the Peripheral Zone using Local Peak Information.
DOI: 10.5220/0004762905100519
In Proceedings of the 3rd International Conference on Pattern Recognition Applications and Methods (ICPRAM-2014), pages 510-519
ISBN: 978-989-758-018-5
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)
the potential to improve the detection of prostate can-
cer in MR spectroscopy. In fact, it also allows detec-
tion of prostate cancer in the transitional zone. On the
other hand, (Ampeliotis et al., 2007) used a combined
feature vectors from MRI T-2 weighted images and
Dynamic Contrast Enhanced (DCE) to increase the
sensitivity of prostate cancer detection. (Engelbrecht
et al., 2010) suggests new techniques such as dynamic
contrast-enhanced MRI, diffusion-weighted imaging,
and magnetic resonance spectroscopic imaging yield
significant improvements in identification and volume
estimation.
In this paper we propose a new method for detect-
ing prostate abnormality within the peripheral zone
based on peak information obtained from the ex-
tracted features. In contrast to the existing methods
in the literature, our method is different in the sense
that:
1. The proposed method does not use any super-
vised/unsupervised classifiers for likelihood clas-
sification of each region as used in (Vos et al.,
2010) and (Ampeliotis et al., 2007).
2. We only used a single modality for abnormal-
ity detection which is T2-Weighted MRI. The
method in (Engelbrecht et al., 2010) used mul-
timodality such as diffusion MRI and MR Spec-
troscopy. Similarly, the method proposed in
(Vos et al., 2010) used a multiparametric MR
of T1- and T2-weighted imaging. (Choi et al.,
2007) suggests that various techniques such as
dynamic contrast materialenhanced MR imag-
ing, diffusion-weighted imaging, and MR spec-
troscopy has the potential to improve the detec-
tion of prostate cancer. On the other hand (Reins-
berg et al., 2007) combined the use of diffusion-
weighted MRI and 1H MR Spectroscopy.
3. The proposed method is purely based on peak in-
formation obtained from extracted features. Un-
like the method in (Vos et al., 2010) they used
additional clinical diagnostic information such as
biopsy tests in making decision as whether cancer
is truly present or not.
2 PROSTATE MODELLING
Figure 2 shows a schematic overview of the prostate.
Approximately 70% of prostate cancer can be found
within the peripheral zone (PZ) and 30% within the
central zone (CZ) and transitional zone (TZ) (Choi
et al., 2007). Therefore, this method aims to de-
tect prostate abnormality only within the PZ. We did
not perform automatic prostate segmentation and the
prostates associated regions were delineated by an ex-
pert radiologist.
Figure 2: CZ = central zone, PZ = peripheral zone, TZ =
transitional zone, T = tumor.
Based on the schematic overview shown in Figure
2, we defined our 2D prostate model in Figure 3. The
prostate model has been recently proposed by (Ram-
pun et al., 2013a). The prostate’s PZ is defined us-
ing the quadratic equation y = ax
2
+ bx + c based on
three crucial coordinate points of the prostate which
are v
1
,v
2
and v
3
. They are determined by the outmost
x and y coordinates of the prostate boundary which are
x
min
,x
max
,y
min
,y
max
(see Figure 3). For example, x
min
and y
max
can be determined by taking the minimum
and maximum x and y coordinates along the prostate
boundary. Moreover, the x coordinates of v
1
and v
2
are captured from x
min
and x
max
and their y coordi-
nate is determined by taking the middle y coordinate
between y
min
and y
max
. On the other hand, the coor-
dinate of v
2
is taken from the middle x coordinate of
x
min
and x
max
and its y coordinate is determined by
taking
7
8
of the distance from y
min
to y
max
. Mathemat-
ically, these can be represented in equations (1), (2),
(3) and (4).
Figure 3: Prostate gland (red) and the defined PZ below
y = ax
2
+ bx + c (green) which goes through v
1
,v
2
and v
3
.
C
p
= ((x
min
+ x
max
)/2,(y
min
+ y
max
)/2) (1)
v
1
= (x
min
,(y
min
+ y
max
)/2) (2)
v
2
= ((x
min
+ x
max
)/2,y
min
+ ((y
max
− y
min
) ×
7
8
))
(3)
v
3
= (x
max
,(y
min
+ y
max
)/2) (4)
where C
p
is the central point of the prostate. Once
the coordinates of v
1
,v
2
and v
3
are defined, we can
DetectionofProstateAbnormalitywithinthePeripheralZoneusingLocalPeakInformation
511
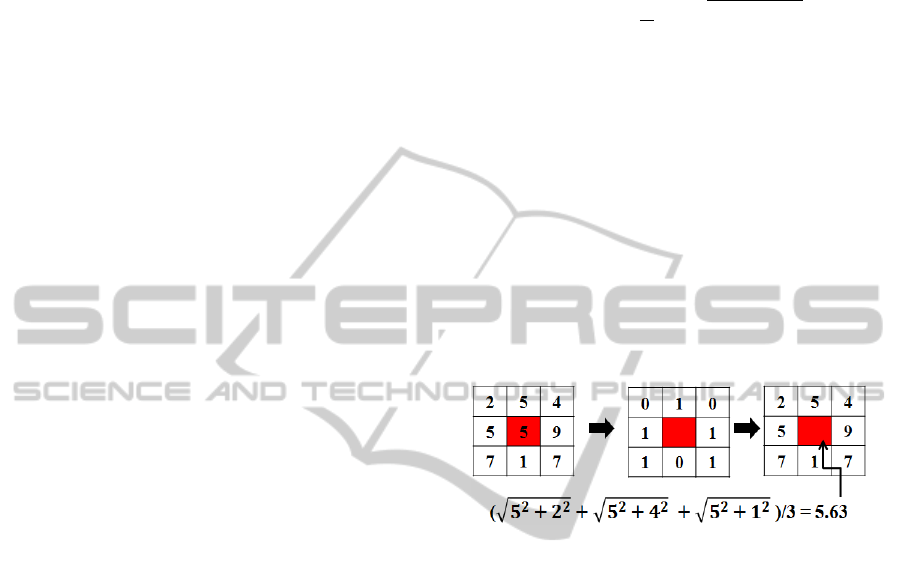
determine the values of a, b and c (therefore a final
quadratic equation is defined). Finally, by taking ev-
ery x coordinate from x
min
to x
max
into quadratic equa-
tion we are able to determine the y coordinate which
will define the boundary of R
1
and R
2
. R
1
(amber re-
gion) and R
2
(turquoise region) are represented by left
and right regions, respectively. By comparing the in-
formation obtained from R
1
and R
2
we estimate the
presence of abnormalities.
3 PROPOSED METHODOLOGY
In summary, the proposed methodology starts by ex-
tracting two features from the original image followed
by smoothing each of the extracted features. Subse-
quently, we performed peak detection within R
1
and
R
2
for both features. Finally, based on the peak infor-
mation obtained, a decision is made as to whether an
abnormality is present.
3.1 Features
In many image analysis techniques, features repre-
senting specific image aspects are important in cap-
turing information representing all textures. This pro-
cess can be done using several techniques such as Ga-
bor Filters (Zheng et al., 2004) (Jain and Karu, 1996),
Grey Level Co-occurence Matrix (GLCM) (Haralick
et al., 1973) (Soh and Tsatsoulis, 1999) (Clausi, 2002)
and Local Binary Pattern (LBP) (Lahdenoja et al.,
2005). According to (Edge et al., 2010) and (Halpern
et al., 2002) most prostate cancers in the PZ tend to
have a dark appearance and several studies suggested
that prostate cancer tissues tend to appear darker on a
T2-weighted MRI image (Garnick et al., 2012) (Ginat
et al., 2009) (Bast et al., 2000). In fact, radiologists
also tend to use darker regions to identify abnormal-
ity within the PZ (Taneja, 2004). In another study
(Vos et al., 2010) showed a potential method to detect
prostate cancer by using blob and peak detectors on
dark blob-like regions within apparent diffusion coef-
ficient (ADC) map. This section presents our features
which are based on low intensities to capture cancer-
ous tissues followed by peak detection which will be
explained in the next section.
In the proposed method, before features are ex-
tracted we performed noise reduction using median
filtering. Median filtering takes the median intensity
value of the pixels within the window as an output in-
tensity of the pixel being processed. We used median
filtering as it can smooth pixels whose values differ
significantly from their surroundings without affect-
ing the other pixels (Ekstrom, 1984). Subsequently,
we extracted two features which are f
low
and f
di f
.
Firstly, f
low
is calculated based on a sum of two vec-
tors (McAllister and Bellittiere, 1996) which can be
computed using
f
low
(p,q) =
1
N
N
∑
b=1
(i
c
)
2
+ (i
b
)
2
(5)
where i
b
is the intensity values of the b
th
neighbour, i
c
is the intensity value of the central pixel and f
low
(p,q)
represents a new intensity value at position (p,q). In
addition, N is the number of neighbours which have
intensity values less than the intensity value of the
central pixel. This means, f
low
represents the average
of the sum vector of the central pixel with each of its
neighbours which have lower intensity value than the
central pixel. f
low
contains most of the low intensities
(higher probobality of cancer). In order to extract the
feature we run a small 5 × 5 sliding window over the
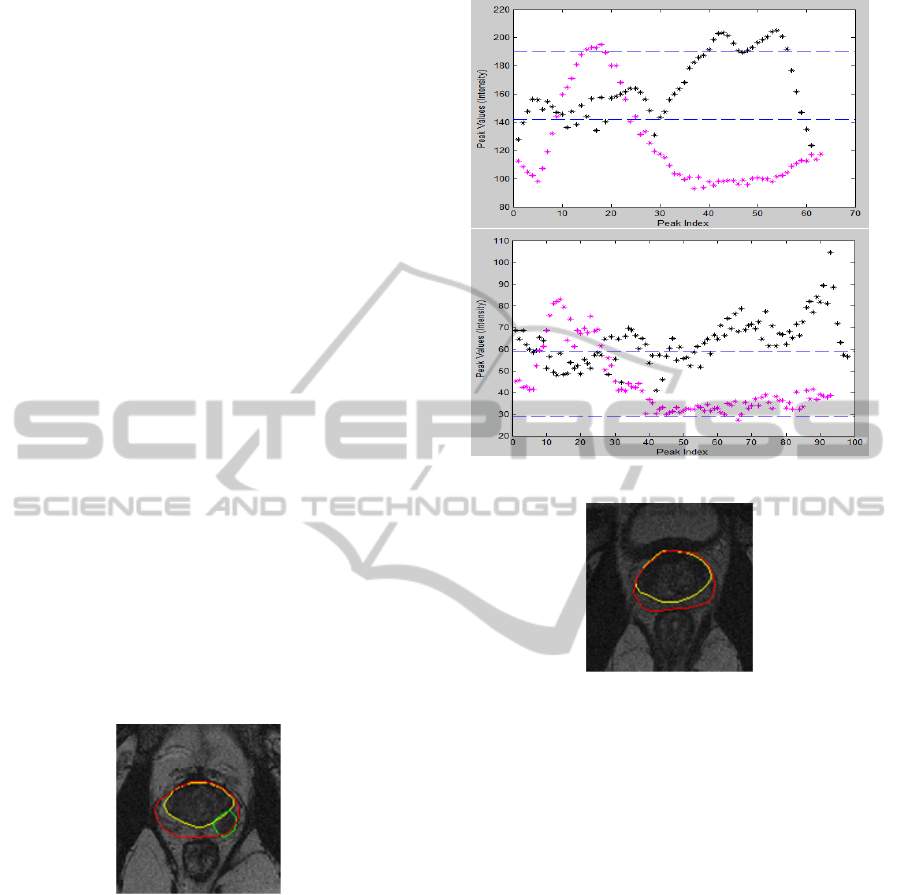
original image (I). Figure 4 shows an example of how
f
low
(p,q) is calculated for a 3 × 3 window.
Figure 4: f
low
(p,q) is calculated using equation (5) by tak-
ing only those neighbours which have a value smaller than
the central pixel.
On the other hand, the second feature ( f
di f
) is cal-
culated by substracting f
min
from each of the elements
in f
low
. This means, f
di f
contains the information
difference between the sum vector and the minimum
value. f
min
can be calculated by taking the minimum
value within a window.
f
min
(p,q) = min
{
g(k, l)
}
(6)
where g is a window, g(k, l) ∈ I, I is the original im-
age and k and l is the dimension of the window used.
Once f
min
is extracted we calculate f
di f
using the fol-
lowing the equation
f
di f
(p,q) = f
low
(p,q) − f
min
(p,q) (7)
This means, f
di f
contains the difference between f
low
and f
min
. f
di f
represents lower intensities than hte
ones in f
low
(this will enhance our chance to capture
more cancerous tissues). For example, based on Fig-
ure 4 f
min
(p,q) = 1 and f
low
(p,q) = 5.63, therefore
f
di f
(p,q) = 4.63. Since we have two features ( f
low
and f
di f
) for each region R
1
and R
2
, from now on we
use R
f
r
1
and R
f
r
2
to //represent R
1
and R
2
for each fea-
ture, where r ∈
{
low,di f
}
. Figure 5 shows examples
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
512

Figure 5: From left is original image, f
low
and f
di f
, respec-
tively.
of features extracted from I. Note that, many black
spots appeared in f
low
. This is due to the effect in
(5) (e.g. if i
c
is smaller than all of its neighbours val-
ues). Since this will affect the peak detection results,
smoothing/noise reduction is necessary to reduce this
problem.
3.2 Feature Smoothing
According to (Hardle, 1991), feature smoothing is a
process to minimise noise in an image while keeping
the most important aspects of a dataset. In the pro-
posed algorithm, we used two-stage noise reduction
(Rampun et al., 2013b) based on a discrete cosine
transform (DCT) followed by replacement of each
pixel by the average of the neighbouring pixel values
(using a 5 × 5 window). The complete explanation
of DCT can be found in (Cabeen and Gent, 2012).
Despite its good performance, DCT does not spec-
ify Q (the amount of smoothing/compression) auto-
matically (Watson, 1993), which mainly affects the
final result of the smoothed image. Garcia’s modi-
fied method (Garcia, 2010) (based on DCT) uses a
smoothing parameter that minimises the generalized
cross-validation (GCV) score to estimate the amount
of smoothing carried out in order to avoid over- or
under-smoothing. Finally, Garcia’s method is robust
in dealing with weighted, missing, and outlien values
by using an iterative procedure (Garcia, 2010). Figure
6 shows an example of f
low
after smoothed.
Figure 6: Smoothed f
low
using DCT. Note that the small
black spots/region (Figure 5) have disappeared.
3.3 Peak Detection
Peak detection is the process of finding local max-
ima and minima of a signal that satisfy certain proper-
ties (Instruments, 2013). Even though peak detection
is less popular in medical image analysis, there are
numerous methods have been developed particularly
in signal processing (Latha et al., 2011). In signal
processing, peak detection is widely used to capture
signal fluctuation by measuring its properties such as
positions, heights and widths. A common way to per-
form this technique is to make use of the fact that the
first derivative of a peak has a downward-going zero-
crossing at the peak maximum. Figure 7 shows an ex-
ample of f
low
within R
1
before and after smoothing.
In the proposed method we applied peak detection in-
Figure 7: Unsmooth and smoothed signal represents in red
and blue line, respectively. Fewer peaks are detected within
the smoothed signal (blue) which is basically reduce the
number of false zero-crossing.
dividually within R
1
and R
2
for each of the features.
Since we are using Ochiai coefficient (Ochiai, 1957)
to measure the similirity between two sets of vectors,
we need to vectorise R
1
and R
2
(
⃗
R
f
r
1
and
⃗
R
f
r
2
, respec-
tively) by taking each element from top to bottom, left
to right. Each element in
⃗
R
f
r
1
will be compared to its
neighboring values. If an element is larger than both
of its neighbors, the element is a local peak (Math-
Works, 2013). Mathematically, a peak is defined as
⃗
R
f
r
1
(p,q) >
⃗
R
f
r
1
(p,q+1) and
⃗
R
f
r
1
(p,q) >
⃗
R
f
r
1
(p,q−1).
This is the same for in
⃗
R
f
r
2
(p,q).
3.4 Abnormality Detection
In the proposed method, abnormality detection is per-
formed by comparing the peak information obtained
from
⃗
R
f
r
1
and
⃗
R
f
r
2
. We calculated the following infor-
mation
1. The Ochiai coefficient(O
f
r
) (Ochiai, 1957) be-
tween
⃗
R
f
r
1
and
⃗
R
f
r
2
which is similar to the cosine
DetectionofProstateAbnormalitywithinthePeripheralZoneusingLocalPeakInformation
513

similarity (Zhu et al., 2010). The Ochiai coeffi-
cient measures the similirity between two sets of
vectors. In principle, any type of similarity co-
efficient can be used, however we chose Ochiai
coefficient because several experiments (Abreu
et al., 2006) (Abreu et al., 2009) have shown that
its performance is better than some other coeffi-
cient (Hofer and Wotawa, 2012) such as Tarantula
(Jones and Harrold, 2005) and Jaccard (Yue and
Clayton, 2005). The Ochiai coefficient indicates,
the higher the value the more similar the elements
in
⃗
R
f
r
1
and
⃗
R
f
r
2
which lead to a lower possibility of
the prostate being abnormal.
2. The percentage (S) of elements in
⃗
R
f
di f
2
that fall
within the range of elements in
⃗
R
f
di f
1
(if
⃗
R
f
di f
2
>
⃗
R
f
di f
1
). The higher the percentage the lower the
difference between
⃗
R
f
di f
1
and
⃗
R
f
di f
2
which means a
lower possibility of the prostate being abnormal.
We will explain how each of the above metrics is cal-
culated in this section. The Ochiai coefficient is de-
fined as
O =
#(A ∩B)
(#(A) ×#(B))
(8)
where A and B are sets which are represented as vec-
tors and #(A) and #(B) are the number of elements in
A and B, respectively. However, since the values in
⃗
R
f
r
1
and
⃗
R
f
r
2
are real values (e.g. 3.123) it is impossi-
ble to calculate #(A ∩ B). On the other hand, we only
take elements in
⃗
R
f
r
n
which are within the minLimit
and maxLimit of each vector. This means, peaks with
values outside the range will be ignored to ensure that
we are considering only realiable peaks and minimise
the effect of noise. We summarise each step for
⃗
R
f
low
n
,
1. Find the range of
⃗
R
f
low
1
and
⃗
R
f
low
2
.
range
n
= max
⃗
R
f
low
n
− min
⃗
R
f
low
n
(9)
where max and min are the maximum and mini-
mum values in
⃗
R
f
low
n
. Assuming that,
⃗
R
f
low
2
has the
smaller range value.
2. Since
⃗
R
f
low
2
has a smaller range, find its minLimit
and maxLimit values (if
⃗
R
f
low
1
has smaller range,
find its minLimit and maxLimit values instead).
minLimit
⃗
R
f
low
2
= µ
⃗
R
f
low
2
− σ
⃗
R
f
low
2
(10)
maxLimit
⃗
R
f
low
2
= µ
⃗
R
f
low
2
+ σ
⃗
R
f
l
ow
2
(11)
µ
⃗
R
f
low
2
=
1
N
N
∑
⃗
R
f
low
2
(p,q) (12)
σ
⃗
R
f
low
2
=
1
N
N
∑
(
⃗
R
f
low
2
(p,q) −µ
⃗
R
f
low
2
) (13)
where µ and σ represent the mean and standard
deviation of
⃗
R
f
low
2
.
3. Calculate the Ochiai coefficient using
O
f
l
ow
=
#(
⃗
R
f
low
1
≤
⃗
R
f
low
2
≤
⃗
R
f
low
1
)
(#(
⃗
R
f
low
1
) ×#(
⃗
R
f
low
2
))
(14)
where #(
⃗
R
f
r
1
≥
⃗
R
f
r
2
≤
⃗
R
f
r
1
) represents the number
of elements in
⃗
R
f
r
1
which is within minLimit
⃗
R
f
r
2
and maxLimit
⃗
R
f
r
2
.
The steps are the same for the second feature ( f
di f
)
in
⃗
R
f
di f
n
. Next, we calculate S (the maximum value is
100) using
S =
#((
⃗
R
f
r
1
≥
⃗
R
f
r
2
≤
⃗
R
f
r
1
)
H
× 100 (15)
where
H =
#
⃗
R
f
di f
1
, i f #
⃗
R
f
di f
1
> #
⃗
R
f
di f
2
#
⃗
R
f
di f
2
, otherwise
(16)
The decision whether abnormality is present or
not is based on the flow chart in Figure 8.
Figure 8: Flow chart decision rule.
Based on the conditions above, it shows that a
slice is malignant if both O
f
low
and O
f
di f
are smaller
than the threshold value (we selected this parame-
ter value as 0.43 as this produced the highest correct
classification rate based on 101 different thresholds
tested (see Figure 14)) or if O
f
low
or O
f
di f
are below
the threshold value and S below 60 (we chose this pa-
rameter value as it produced the highest accuracy of
≈ 70% based on 10 different thresholds (10 to 100)
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
514
tested. The experiment was performed by classifying
every single case using a single S value and selecting
the one with the highest classification rate. This ex-
periment is exactly the same when selecting the value
of O
f
low
and O
f
di f
). This means, if any of O
f
low
or
O
f
di f
value is smaller than the threshold, we will use
S to determine the occurance of abnormalities. Nev-
ertheless, if S = 100, we do not have a need to check
O
f
r
because it indicates the maximum similarity per-
centage. Finally, if the third condition in Figure 8 is
passed then the slice is considered benign.
4 EXPERIMENTAL RESULTS
This section presents our experimental results. In to-
tal, our database contains 90 prostate T2-Weighted
MRI images (512 × 512) from 25 different patients
aged 54 to 74. Each patient has 3 to 5 slices through
the central part of the prostate. The prostates, cancer
and central zones were delineated by an expert radi-
ologist on each of the MRI images. Data was anal-
ysed and classified as to whether the prostate con-
tains cancer based on the conditions explained pre-
viously. Subsequently, we compared the result with
the ground truth whether the prostate contains cancer
regions or not. We will present two different cases
and the justification for the threshold values selec-
tion. The first case (Figure 9) shows the results of
a slice with cancer. Figure 9 shows the original image
Figure 9: Case # 1: Malignant.
(top) with ground truth, prostate gland (red), CZ (yel-
low) and tumor (green). Figure 10 shows results of
⃗
R
f
low
n
and
⃗
R
f
di f
n
, respectively. Black and magenta rep-
resent
⃗
R
f
r
1
and
⃗
R
f
r
2
. On the other hand, maxLimit
⃗
R
f
r
n
and minLimit
⃗
R
f
r
n
are represented in the lines which are
parallel with the x axis (broken lines). Note that we
use the same order and notations for the second case
as well. From Figure 10 we can visually identify that
most values in
⃗
R
f
r
1
and
⃗
R
f
r
2
are quite separated in both
features. By calculating the matrics defined in section
3.4, we will get the following results: O
f
low
= 0.177,
O
f
di f
= 0.367 and S = 35.71%. Therefore the slice is
considered malignant based on the conditions shown
Figure 10: Results of case # 1: Malignant.
Figure 11: Case # 2: Benign.
in section 3.4.
On the other hand, Figures 11 and 12 show an ex-
ample of a slice without an abnormality and its re-
sults, respectively. In this example, we can see that
≈ 95% of the values in
⃗
R
f
r
1
and
⃗
R
f
r
2
are within the same
range (Figure 12). By calculating the metrics we will
get the following results: O
f
low
= 0.77, O
f
di f
= 0.81
and S = 72.44%. Therefore, the slice is considered
benign because all metric values are above the thresh-
old values.
Figure 13 shows the Receiver Operating Charac-
teristic (ROC) curves of the proposed method based
on 101 different threshold values. The minimum and
maximum threshold values are 0 and 1, respectively
with 0.01 difference between threshold values. The
ROC graph shows that the proposed method achieved
0.80 true positive rate with 0.20 false positive rate.
In terms of correct classification rate (CCR) against
different thresholds, Figure 14 shows that O
f
r
< 0.43
achieved the highest CCR of 82.2%. On the other
hand, Figure 14 shows the boxplot of O
f
low
and O
f
di f
for benign (blue) and malignant (red) cases. Based on
the results in Figure 14, it is clearly shows that most
DetectionofProstateAbnormalitywithinthePeripheralZoneusingLocalPeakInformation
515
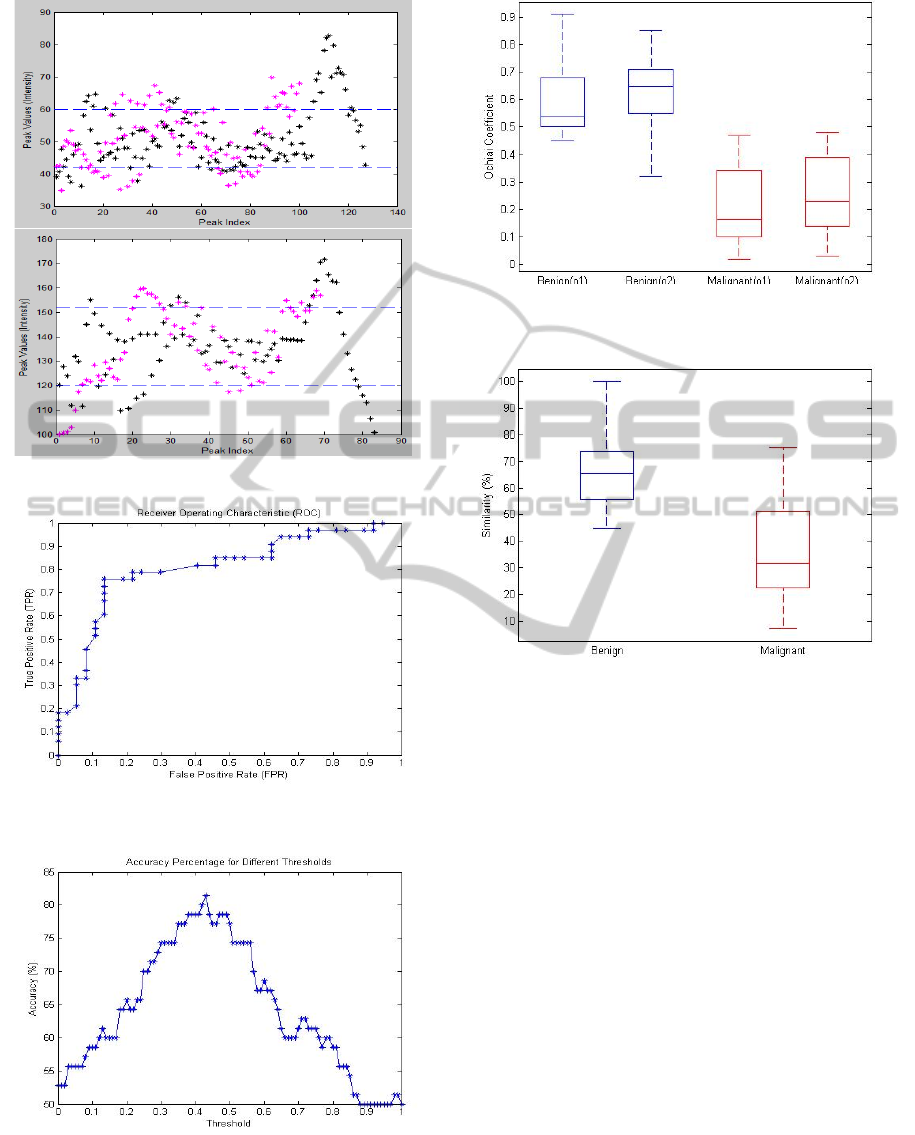
Figure 12: Results of case # 2: Benign.
Figure 13: ROC curves of the proposed method using 101
different threshold values (O
f
r
).
Figure 14: Accuracies of the proposed method using 101
different threshold values (O
f
r
).
benign cases have O
f
r
> 0.4 between
⃗
R
f
r
1
and
⃗
R
f
r
2
.
Similarly the results in Figure 15 shows that most be-
Figure 15: O
f
l ow
and O
f
di f
represented in o1 and o2, respec-
tively.
Figure 16: Similarity percentage (S) (between
⃗
R
f
r
1
and
⃗
R
f
r
2
)
for benign and malignant cases represented in blue and red,
respectively.
nign cases have S > 60 compared to malignant cases
which have S < 60.
For the 90 prostate MRI slices from 25 different
patients (41 slices are malignant and 49 slices are be-
nign), the proposed method achieved 82.2% (74 slices
classified correctly). On the other hand, the proposed
method produced 8.9% false negative and false posi-
tive results. Since, the number of methods which uses
peak detection methods (in medical image analysis
applications) in the literature is limited, it is hard to
make a direct comparison with other methods. In ad-
dition, different datasets and frameworks used in the
literature also make it extremely difficult to perform
a qualitative comparison. However, to compare our
results indirectly with some existing methods we cite
several methods which have a similar goal (detecting
prostate abnormality/cancer).
Table 1 presents the experimental results of ten
different methods (included our method) and their
average accuracies. Note that every method used
different datasets, modalities and frameworks. The
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
516

Table 1: First, second and third column represents the au-
thors, number of prostates and average accuracy (AA), re-
spectively.
Authors # of cases AA (%)
(Sung et al., 2011) 42 89
(Vos et al., 2010) 29 89
(Reinsberg et al., 2007) 42 87
(Rampun et al., 2013a) 19 85
(Ampeliotis et al., 2007) 10 84
Our method 25 82
(Castaneda et al., 2009) 15 80
(Litjens et al., 2011) 188 79
(Tiwari et al., 2007) 14 78
(Llobet et al., 2007) 303 62
methods proposed by (Sung et al., 2011) and (Vos
et al., 2010) achieved the highest average accuracy of
89% whereas the methods by (Llobet et al., 2007),
(Tiwari et al., 2007), (Litjens et al., 2011) failed
to achieve more than 80% accuracy. On the other
hand, the proposed method achieved 0.81 sensitiv-
ity and 0.84 specificity which is similar with the re-
cent method proposed by (Artan and Yetik, 2012).
(Girouin et al., 2007) reports sensitivity/specificity
value of 0.5-0.6 and 0.13-0.21 over 46 patients us-
ing only T2-weighted (1.5 T) MRI. Further, (Girouin
et al., 2007) presents a higher both sensitivity and
specificity 0.83 for the same number of patients us-
ing T2-weighted (3.0 T) MRI. In a smaller dataset
(Ftterer et al., 2006) presents 0.83 both on sensitivity
and specificity, respectively. In a different modality,
(Wong and Scharcanski, 2011) shows a higher sen-
sitivity (0.93) and specificity (0.96) in 46 ultrasound
images. As mentioned earlier, these comparisions are
very subjective as accuracy, sensitivity and specificity
are highly influenced by the number of datasets, dif-
ferent modalities and methods’ frameworks. There-
fore it is extremely difficult to make a real compar-
ison either quantitatively or qualitatively. Some ob-
vious drawbacks of the proposed method, it failed to
produce accurate results in two cases: a) when the
prostate’s peripheral zone is almost non-existent, and
b) when the prostate’s shape does not conform to the
shape of our prostate model.
5 CONCLUSIONS
The proposed method shows that peak information
has a potential role in detecting prostate abnormalities
within the PZ. In contrast with some of the methods
in Table 1, our method uses a minimal set of features
and modalities (only T2-Weighted MRI). Methods by
(Engelbrecht et al., 2010) and (Vos et al., 2010) used
multimodalities data such as diffusion MRI, MR spec-
troscopy and a combination of T1 and T2-weighted
imaging. Secondly, the proposed method does not
use any supervised/unsupervised classifiers for clas-
sification but it is entirely dependent on the metric
values computed from the peak information. More-
over, our method does not refer to or use any clinical
features in making decision as to whether abnormal-
ity occurs but is entirely based on the values of O
f
r
and S. The method of (Vos et al., 2010) used clinical
features such as PSA level to support their method in
making decision whether abnormality is truly present
or not. Nevertheless, one obvious drawback of our
method is that it does not have an additional method to
reduce false positive and false negative results. There-
fore a robust false positive/negative reduction method
is needed to increase the accuracy rate. Secondly, if
the tumor appears outside the R
1
and R
2
regions the
method failed to identify the abnormality due to the
incorrect information captured. To solve this issue,
we are developing a segmentation method to sepa-
rate CZ from prostate gland. This allows the proposed
method to be more robust because it will analyse re-
gions within the prostate (therefore improving the 2D
PZ model will be apart of our future work).
In short, with 8.9% false negative and false pos-
itive results (sensitivity/specificity: 0.81/0.84), the
proposed method achieved similar accuracy with
some of the methods in the literature. In our study, we
have shown that most MRI slices containing cancer
have small value of Ochiai coefficient (O
f
r
< 0.43)
and S < 60%. This means, if the coefficient be-
tween
⃗
R
f
r
1
and
⃗
R
f
r
2
is less than a threshold value (in
our case 0.43), the prostate has a higher probability
to be abnormal. Moreover, a similarity percentage
less than 60% also indicate prostate abnormality. Fi-
nally, the next stage of this research is to test the pro-
posed method on a larger dataset with a combination
of several methods (e.g. CZ segmentation, false pos-
itive/negative reduction, and blob detection), improv-
ing the 2D PZ model and other statistical features (e.g.
entropy and energy).
REFERENCES
Abreu, R., Zoeteweij, P., Golsteijn, R., and van Gemund,
A. J. (2009). A practical evaluation of spectrum-based
fault localization. Journal of Systems and Software,
82(11):1780 – 1792. ¡ce:title¿SI: {TAIC} {PART}
2007 and {MUTATION} 2007¡/ce:title¿.
Abreu, R., Zoeteweij, P., and Van Gemund, A. J. C. (2006).
An evaluation of similarity coefficients for software
fault localization. In Dependable Computing, 2006.
DetectionofProstateAbnormalitywithinthePeripheralZoneusingLocalPeakInformation
517

PRDC ’06. 12th Pacific Rim International Symposium
on, pages 39–46.
Ampeliotis, D., Antonakoudi, A., Berberidis, K., and
Psarakis, E. Z. (2007). Computer aided detection of
prostate cancer using fused information from dynamic
contrast enchanced and morphological magnetic res-
onance images. In IEEE International Conference
on Signal Processing and Communications(ICSPC
2007).
Artan, Y. and Yetik, I. (2012). Prostate cancer localiza-
tion using multiparametric mri based on semisuper-
vised techniques with automated seed initialization.
Information Technology in Biomedicine, IEEE Trans-
actions on, 16(6):1313–1323.
Aus, G., Hermansson, C. G., Hugosson, J., and Pedersen,
K. V. (2008). Transrectal ultrasound examination of
the prostate: complications and acceptance by pa-
tients. British journal of urology, 71(4):457–459.
Bast, R. C., Kufe, D. W., Pollock, R. E., Weichselbaum,
R. R., Holland, J. F., Frei, E., Halvorsen, R. A., and
Thompson, W. M. (2000). Imaging neoplasms of the
abdomen and pelvis.
Brawer, M. K. (1991). Prostate specic antigen: A review.
Acta Oncologica., 30(2):161–168.
Cabeen, K. and Gent, P. (2012). Image com-
pression and the Discrete Cosine Transform.
http://www.lokminglui.com/dct.pdf/. Accessed
11-August-2013.
Castaneda, B., An, L., Wu, S., Baxter, L. L., Yao, J. L.,
Joseph, J. V., Hoyt, K., Strang, J., Rubens, D., and
Parker, K. J. (2009). Prostate cancer detection using
crawling wave sonoelastography. In Proc. SPIE 7265,
Medical Imaging 2009: Ultrasonic Imaging and Sig-
nal Processing.
Choi, Y. F., Kim, F. K., Kim, N., Kim, K. W., Choi, E. K.,
and Cho, K.-S. (2007). Functional MR imaging of
prostate cancer. RadioGraphics, 27(1):63–68.
Clausi, D. A. (2002). An analysis of co-occurrence texture
statistics as a function of grey level quantization. Can.
J. Remote Sensing, 28(1):45–62.
Edge, S. B., Byrd, D. R., Compton, C., Fritz, A. G., Greene,
F. L., and Trotti, A. (2010). AJCC Cancer Staging
Manual (7th Edition). Springer., Chicago, US.
Ekstrom, P. M. (1984). Digital Image Processing Tech-
niques. Academic Press, Inc., London, UK.
Engelbrecht, M. R., Puech, P., Colin, P., Akin, O. Lemaitre,
L., and Villers, A. (2010). Multimodality magnetic
resonance imaging of prostate cancer. Journal of en-
dourology society, 24(5):677–684.
Ftterer, J. J., Heijmink, S. W. T. P. J., Scheenen, T. W. J.,
Veltman, J., Huisman, H. J., Vos, P., de Kaa, C.
A. H., Witjes, J. A., Krabbe, P. F. M., Heerschap,
A., and Barentsz, J. O. (2006). Prostate cancer lo-
calization with dynamic contrast-enhanced mr imag-
ing and proton mr spectroscopic imaging. Radiology,
241(2):449–458. PMID: 16966484.
Garcia, D. (2010). Robust smoothing of gridded data in one
and higher dimensions with missing values. Computa-
tional Statistics and Data Analysis, 54(4):1167–1178.
Garnick, M. B., MacDonald, A., Glass, R., and Leighton,
S. (2012). Harvard Medical School 2012 Annual Re-
port on Prostate Diseases. Harvard Medical School,
Harvard , US.
Ginat, D. T., Destounis, S. V., Barr, R. G., Castaneda, B.,
Strang, J. G., and Rubens, D. J. (2009). Us elastog-
raphy of breast and prostate lesions1. Radiographics,
29(7):2007–2016.
Girouin, N., M
`
ege-Lechevallier, F., Senes, A. T., Bissery,
A., Rabilloud, M., Mar
´
echal, J., Colombel, M., Ly-
onnet, D., and Rouvi
`
ere, O. (2007). Prostate dy-
namic contrast-enhanced mri with simple visual diag-
nostic criteria: is it reasonable? European radiology,
17(6):1498–1509.
Halpern, E. J., Cochlin, D. L., and Goldberg, B. (2002).
Imaging of the Prostate. Martin Dunitz Ltd., London,
UK.
Haralick, R. M., Shanmugam, K., and Dinstein, I. (1973).
Textural features for image classification. IEEE
Transactions On Systems, Man, and Cybernetics,
3(6):610–621.
Hardle, W. (1991). Smoothing Techniques With Implemen-
tation in S. Springer-Verlag., Louvain-La-Neuve, Bel-
gium.
Hofer, B. and Wotawa, F. (2012). Spectrum enhanced dy-
namic slicing for better fault localization. In ECAI,
pages 420–425.
Instruments (2013). Wavelet-based peak detection.
http://www.ni.com/white-paper/5432/en/. Accessed
24-July-2013.
Jain, A. K. and Karu, K. (1996). Learning texture discrim-
ination. IEEE Transactions on Pattern Analysis and
Machine Intelligence, 18:195–205.
Jones, J. A. and Harrold, M. J. (2005). Empirical evaluation
of the tarantula automatic fault-localization technique.
In Proceedings of the 20th IEEE/ACM International
Conference on Automated Software Engineering, ASE
’05, pages 273–282, New York, NY, USA. ACM.
Kenny, T. (2012). Prostate cancer.
http://www.patient.co.uk/health/prostate-cancer/.
Accessed 12-August-2013.
Lahdenoja, O., Laiho, M., and Paasio, A. (2005). Local bi-
nary pattern feature vector extraction with cnn. In 9th
International Workshop on Cellular Neural Networks
and Their Applications, pages 202–205. IEEE Cat.
Latha, I., Reichenbach, S. E., and Tao, Q. (2011). Compar-
ative analysis of peak-detection techniques for com-
prehensive two-dimensional chromatography. Radio-
Graphics, 1218(38):6792–6798.
Litjens, G. J. S., Vos, P. C., Barentsz, J. O., Karssemeijer,
N., and Huisman, H. J. (2011). Automatic computer
aided detection of abnormalities in Multi-Parametric
prostate MRI. In Proc. SPIE 7963, Medical Imaging
2011: Computer-Aided Diagnosis.
Llobet, R., Juan, C., Cortes, P., Juan, A., and Toselli, A.
(2007). Computer-aided detection of prostate can-
cer. International Journal of Medical Informatics,
76(7):547–556.
MathWorks (2013). Documentation center: findpeaks.
ICPRAM2014-InternationalConferenceonPatternRecognitionApplicationsandMethods
518

http://www.mathworks.co.uk/help/signal/ref/findpeaks.html/.
Accessed 23-August-2013.
McAllister, H. C. and Bellittiere, D.
(1996). The sum of two vectors.
http://www.hawaii.edu/suremath/jsumTwoVectors.html.
Accessed 19-August-2013.
Ochiai, A. (1957). Zoo geographical studies on the solenoid
fishes found Japan and its neighboring regions.ii.
Bull.Japan.Soc.Sci.Fisheries., 22(9):526–530.
PCUK (2013). Prostate cancer facts and figures.
http://prostatecanceruk.org/information/prostate-
cancer-facts-and-figures/. Accessed 11-August-2013.
Rampun, A., Chen, Z., and Zwiggelaar, R. (2013a). Detec-
tion and localisation of prostate abnormalities. In 3rd
International Conference on Computational & Math-
ematical Biomedical Engineering, CMBE 13.
Rampun, A., Strange, H., and Zwiggelaar, R. (2013b).
Texture segmentation using different orientations of
GLCM features. In 6th International Conference on
Computer Vision / Computer Graphics Collaboration
Techniques and Applications, MIRAGE 13.
Reinsberg, S. A., Payne, G. S., Riches, S. F., Ashley,
S., Brewster, J. M., Morgan, V. A., and deSouza,
N. M. (2007). Combined use of Diffusion-Weighted
MRI and 1H MR Spectroscopy to increase accuracy
in prostate cancer detection. American Journal of
Roentgenology, 188(1):1122–1129.
Roehl, K. A., Antenor, J. A. V., and Catalona, W. J.
(2002). Serial biopsy results in prostate cancer screen-
ing study. British journal of urology, 167(6):2435–
2439.
Shirley, A. and Brewster, S. (2011). Expert review: The
digital rectal examination. Journal of Clinical Exami-
nation, 11:1–12.
Soh, L. and Tsatsoulis, C. (1999). Texture analysis of sar
sea ice imagery using gray level co-occurrence matri-
ces. Geoscience and Remote Sensing, IEEE Transac-
tions on, 37(2):780–795.
Sung, Y. S., Kwon, H.-J., Park, B. W., Cho, G., Lee,
C. K., Cho, K.-S., and Kim, J. K. (2011). Prostate
cancer detection on dynamic contrast-enhanced mri:
Computer-aided diagnosis versus single perfusion pa-
rameter maps. American Journal of Roentgenology,
197(5):1122–1129.
Taneja, S. S. (2004). Imaging in the diagnosis and manage-
ment of prostate cancer. Reviews in Urology, 6(3):101.
Tempany, C. and Franco, F. (2012). Prostate MRI: Update
and current roles. Applied Radiology, 41(3):17–22.
Tidy, C. (2013). Prostate specific antigen (PSA) test.
http://www.patient.co.uk/health/prostate-specific-
antigen-psa-test/. Accessed 12-August-2013.
Tiwari, P., Madabhushi, A., and Rosen, M. (2007). A hier-
archical unsupervised spectral clustering scheme for
detection of prostate cancer from magnetic resonance
spectroscopy (MRS). In Medical Image Computing
and Computer Assisted Intervention (MICCAI), pages
278–286.
Vos, P. C., Hambrock, T., Barentsz, J., and Huisman, H.
(2010). Computer-assisted analysis of peripheral zone
prostate lesions using T2-weighted and dynamic con-
trast enhanced T1-weighted MRI. Physics in Medicine
and Biology, 55:1719–1734.
Watson, A. B. (1993). Dctune: A technique for visual op-
timization of dct quantization matrices for individual
images. Society for Information Display Digest of
Technical Papers, XXIV:946–949.
Wong, A. and Scharcanski, J. (2011). Fisher #x2013;tippett
region-merging approach to transrectal ultrasound
prostate lesion segmentation. Information Technology
in Biomedicine, IEEE Transactions on , 15(6):900–
907.
Yue, J. C. and Clayton, M. K. (2005). A similarity mea-
sure based on species proportions. Communications in
Statistics - Theory and Methods, 34(11):2123–2131.
Zheng, D., Zhao, Y., and Wang, J. (2004). Feature extrac-
tion using a gabor filter family. In Proceedings of
the 6th IASTED International Conference on Signal
and Image Processing. The International Association
of Science and Technology for Development.
Zhu, S., Wu, J., and Xia, G. (2010). Top-k cosine similar-
ity interesting pairs search. In Seventh International
Conference on Fuzzy Systems and Knowledge Discov-
ery (FSKD).
DetectionofProstateAbnormalitywithinthePeripheralZoneusingLocalPeakInformation
519
