
MongoDB for Electrophysiology Experiments
Petr Je
ˇ
zek, Roman Mou
ˇ
cek and Jakub Dan
ˇ
ek
New Technologies for the Information Society, Department of Computer Science and Engineering,
Faculty of Applied Sciences, University of West Bohemia, Univerzitn
´
ı 8, 306 14 Plze
ˇ
n, Czech Republic
Keywords:
Data, Metadata, Experiment, EEG, ERP, EEG/ERP Portal, Oracle, Mongo, Performance, Flexibility, Fixed-
schema, Free-schema.
Abstract:
Many efforts are devoted to provide a unified solution for maintaining data from electrophysiological experi-
ments. Because large data collections of heterogeneous nature are obtained, neuroinformatics databases must
be robust and flexible. Current database systems are of two types. The first one uses a fixed schema while the
second one is schema free. This paper discusses usage of a NoSQL database, MongoDB, for electrophysiolog-
ical experiments and investigates transformation of existing electroencephalography (EEG) and event-related
potentials (ERP) database records in Oracle into MongoDB. Two perspectives, flexibility and performance are
discussed. A final approach that profits from combination of both concepts, is also discussed.
1 INTRODUCTION
Since laboratories performing electrophysiological
experiments are facing a lot of difficulties with data
storing, management and sharing, researchers are
consistently working on definition of data standards
for the electrophysiological data. The main scientific
initiatives lead not only in definition of standards for
raw data, but more significantly, in definition of stan-
dards for metadata. The ability to deal with experi-
mental metadata significantly helps with better under-
standing of experimental raw data. These difficulties
are solved within Neuroinformatics Coordinating Fa-
cility (INCF) that released recommendations (van Pelt
and van Horn, 2007) for handling of the experimental
data/matadata.
Our research group specializes in the research of
brain activity using methods of electroencephalogra-
phy (EEG) and event related potentials (ERP). As
a member of Czech INCF National Node (CNNN)
we cooperate in definition of data standards for the
EEG/ERP domain.
A large amount of obtained data (tens megabytes
per experiment) and metadata are stored in a custom
system - the EEG/ERP Portal (Jezek and Moucek,
2012) developed as a part of the CNNN infrastructure
(Jezek et al., 2013). It is a mature web based tool de-
veloped to help fulfil INCF goals in standardization of
data formats in electrophysiology. Currently, the data
layer of the EEG/ERP Portal is build on Oracle re-
lational database management system (RDBMS). Al-
though this RDBMS ensures robustness of the system
its concept based on a relational model causes inflex-
ibility. The aim of this paper is to investigate possi-
bilities to migrate records from a relational database
with a fixed schema to a database with a free schema.
This approach is investigated from performance and
flexibility perspectives.
The remainder of this paper is organized as fol-
lows. We start with a review of the state-of-the-art,
pointing out limitations of current approaches. Then,
we briefly introduce the most important part of the
data model in the EEG/ERP Portal. For the reasons
mentioned above a part of this paper is dedicated to
explore whether and how NoSQL databases could
be used as a persistence layer of the EEG/ERP Por-
tal. The last part before conclusion introduces per-
formance testing done to discuss comparison of a re-
lational database in contrast with a NoSQL database.
Finally, the following questions are discussed: What
changes to the application would have to be done to
change the database engines? What disadvantages
would the change brings?.
2 STATE OF THE ART
Several databases for electrophysiological data are
gradually formed. (Rautenberg et al., 2011) presents
a data management system for electrophysiological
422
Ježek P., Mou
ˇ
cek R. and Dan
ˇ
ek J..
MongoDB for Electrophysiology Experiments.
DOI: 10.5220/0004903604220427
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2014), pages 422-427
ISBN: 978-989-758-010-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

data based on a well established relational database
technology (PostgreSQL) together with mechanism
to deal with the heterogeneity of electrophysiologi-
cal data. Different data models link by specific ”glue”
tables. It enables management heterogeneous data.
INCF Dataspace (Group, 2013) is a solution pro-
vided by INCF. It associates INCF nodes data sources
in a distributed system based on iRods solution. Tech-
nically, data are managed locally by individual nodes
and connected using catalogue servers. From a user
perspective it works as a large data file system ac-
cessed through a web interface.
Although this solution is flexible, when such data
are intended to be shared among different systems,
metadata must be provided in a unified format.
Open MetaData Markup Language (OdML)
(Grewe et al., 2011) is a flexible data format introduc-
ing a tree structure of Sections and Properties. The
Property is a core entity of the odML data model that
can contain values. Properties are logically grouped
in sections that can consists of subsections.
Hierarchical Data Format (HDF5) (The HDF
Group, 2010) is a self-describing data format de-
signed to store and organize large amounts of numer-
ical data. The file structure is based on two major
types of objects: Datasets (multidimensional arrays
of a homogeneous type), and Groups (container struc-
tures which can hold datasets and other groups).
StorageBIT (Carreiras et al., 2013) is a solution
that combines HDF5 format (that ensures data persis-
tency) with MongoDB used as a front end for data
access.
Due to heterogeneous nature of data, a growing
number of developers and users have begun turning
to various types of non relational databases (Leav-
itt, 2010). The existence of a broad range of NoSQL
databases on the one hand and efforts to provide flex-
ible data formats on the other hand was the motiva-
tion to investigate replacement of common relational
database by their NoSQL alternatives. Our EEG/ERP
Portal served as a suitable test case. Our tests simul-
taneously validated a StorageBIT approach.
3 EEG/ERP PORTAL DATA
STRUCTURE
The internal structure of the EEG/ERP Portal pro-
vides a set of metadata organized in several seman-
tic groups. The core structure contains the following
metadata:
• Scenario of experiment (name, length, descrip-
tion, ...)
• Experimenters, testing persons, experiment owner
(given name, surname, contact, experiences,
handicaps, ...)
• Description of raw data (format, sampling fre-
quency, ...)
• Codebooks (weather, environment notes, elec-
trode settings, artifact information, ...)
4 IMPLEMENTATION
4.1 Metadata Selection
Because of the EEG/ERP Portal database is extensive
a proof-of-concept working on a restricted subset of
metadata is presented. The set of selected metadata
represents a typical subset of items most often used
when data are queried:
• start and end time of an individual experiment
• research group the experiment belongs to includ-
ing its name and owner’s name
• name of the experiment owner (the responsible
person)
• experiment scenario including its title, description
and name of the scenario’s owner
• subject information including name, date of birth,
laterality, gender and age
• codebooks including surrounding weather, envi-
ronment notes, electrode settings and artifact in-
formation
4.2 Data Models
The database shown in Figure 1 represents all meta-
data selected in Section 4.1.
Because MongoDB represents data in a BSON
format
1
, a BSON document representing the data
structure from Figure 1 has been designed (Listing 1).
Listing 1: JSON Schema that is equivalent to the database
structure. Individual values are demonstration examples.
They do not represent real persons or real experiments.
{
" ex pe ri me nt _i d " : 1,
" h e a der " : {
" s t a r t _ t i m e " : " s tar t t ime " ,
" e n d _ t ime " : " e nd t ime " ,
" o w ner " : {
" f i r s t n a me " : " O liv e r " ,
" l a s t n ame " : " T w ist "
},
" re s e a r c h _ g r o u p " : {
1
BSON is a binary representation of JSON documents,
though contains more data types than does JSON
MongoDBforElectrophysiologyExperiments
423
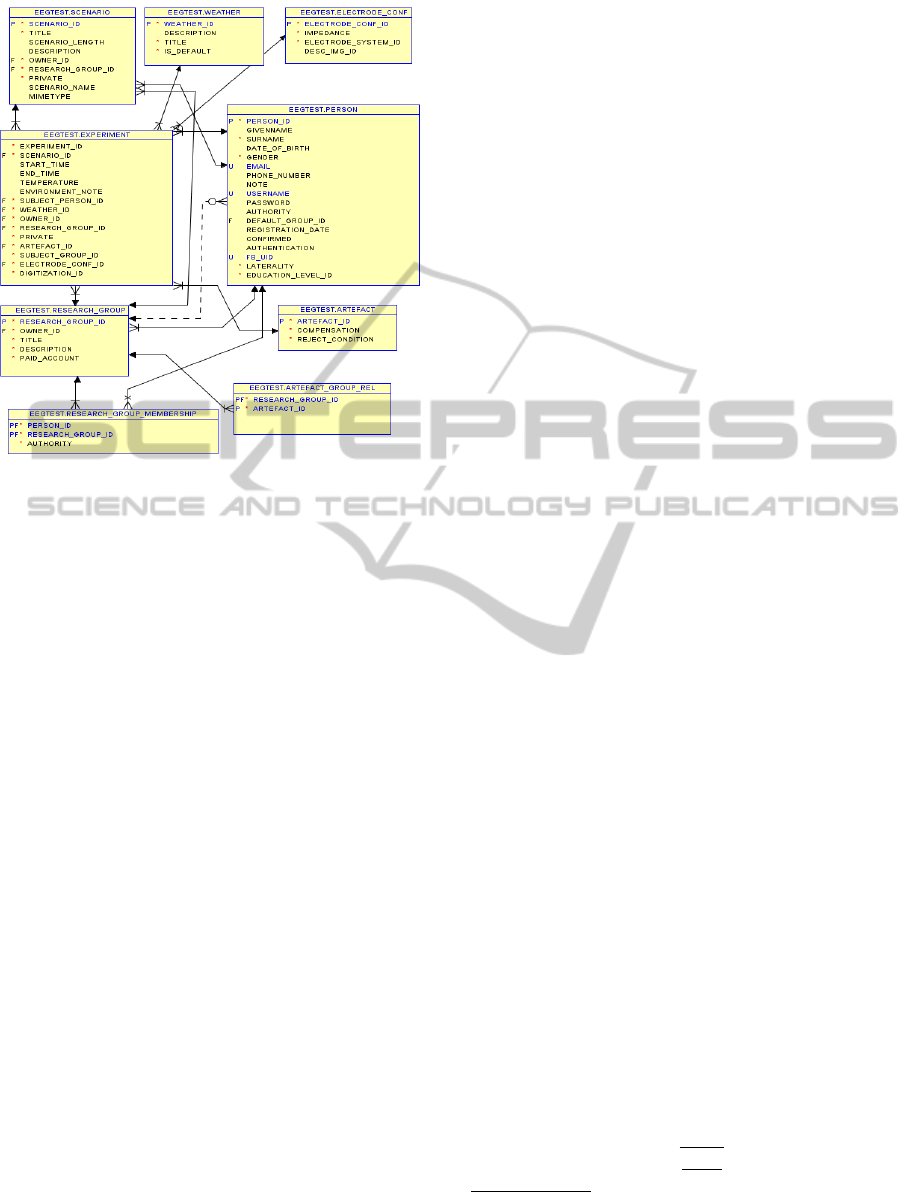
Figure 1: ERA model containing representative subset of
stored metadata.
" t i tle " : " EE G / E RP G r o up " ,
" o w ner " : {
" f i r s t n a me " : " Joh n " ,
" l a s t n ame " : " S m ith "
}
}
},
" s c e n a rio " : {
" n ame " : " dr iv er ’s at t e n tio n " ,
" de s c r i p t i o n " : " Dri ve r ’ s at t e n t i on - a u d i t ory
st i m u l a t i o n of d r iv e r a nd pa s s e nger " ,
" o w ner " : {
" f i r s t n a me " : " O liv i a " ,
" l a s t n ame " : " D u n ham "
}
},
" s u b j ect " : {
" f i r s t n a me " : " Jac k " ,
" l a s t n ame " : " B l ack " ,
" g e n der " : " g e nde r " ,
" d ate of b i rth " : "0 8 /0 1 / 1 9 8 8 " ,
" l a t e r a l i t y " : " r igh t h and " ,
" g r oup " : {
" t i tle " : " EE G / E RP G r o up " ,
" d e s c r i p t i o n " : " A g ro u p e s t a b l i s h ed to p erf o r m
EE G / ER P e x p e r i m e nts "
}
},
" co n f i g u r a t i o n " : {
" te m p e r a t u r e " : 2 2 ,
" w e a t her " : {
" t i tle " : " S unn y " ,
" d e s c r i p t i o n " : " S unn y w e a t her "
},
" en v i r o n m e n t _ n o t e " : " It c o n t r ibut e t o a b ett e r
mo od of te st p er s o n s " ,
" a r t e f act " : {
" co m p e n s a t i o n " : " u se d f i l ter th at re m o ves 5 0 Hz
fr e q u e n cy " ,
" re j e c t _ c o n d i t i o n " : " it doe s not w ea r the
si g n al "
},
" el e c t r o d e _ s y s t e m " : "1 0 - 2 0 sy s tem " ,
" di g i t i z a t i o n " : {
" sa m p l i n g _ r a t e " : "1 0 2 4 " ,
" g ain " : "1 . 2 "
}
}
}
4.3 Test Framework
A test framework has been developed for easier and
repeatable testing
2
. It is written in Python which
has been chosen because contains drivers for both the
database engines, provides good handling of data col-
lections and enables fast development. The frame-
work provides the following functionality:
• Parameter-based generation of test data into Ora-
cle DB
• Export of all experiments from Oracle DB into
MongoDB
• Running bulks of queries on both databases.
It enables to measure execution time, define
parameter-based bulk size or run bulks in paral-
lel to increase server load.
4.4 Test Cases
The tests were run on a Gentoo machine (kernel:
3.7.10, Intel i7 with 4 cores, 8GB of RAM), with
the necessary code implemented in Python. Oracle
database (11g Release 2) and MongoDB 2.4.0. were
used.
We defined three test cases used to compare Mon-
goDB with Oracle performance:
• Search by scenario title (single join in Oracle) -
Test Case 1
• Search by title of a research group’s owner (two
joins in Oracle) - Test Case 2
• Search by age of a subject (aggregation function
in Oracle, date comparison in MongoDB) - Test
Case 3
5 RESULTS
5.1 Performance
Each test consisted of 100 queries in 6 iterations (N).
The tests were run in 1, 2, 4, 8, 16, and 32 paral-
lel processes in each iteration. Therefore 100, 200,
400, 800, 1600 and 3200 queries had been run in each
iteration. The performance (P) was measured in op-
erations per second (op/s) by averaging (avg(s)) how
long each iteration took to run (t
i
) (Equation 1).
P =
op
∑
N
i=1
t
i
N
(1)
2
availabe from: https://github.com/NEUROINFOR
MATICS-GROUP-FAV-KIV-ZCU/EEG-ERP-Portal-
Mongo-PoC
HEALTHINF2014-InternationalConferenceonHealthInformatics
424

Detailed results can be found in Table 1. The ta-
ble is split into two parts (the left part for Oracle, the
right part for MongoDB). For MongoDB six itera-
tions were executed without indexes (the upper half
of table), followed by six iterations with indexes cre-
ated (the bottom part of table). For Oracle, all 12 it-
erations were executed with indexes created for the
relevant columns. For simplicity, results for individ-
ual iterations are omitted, table shows only averaged
results over all iterations.
From Table 1 we can observe:
• MongoDB returns results on average 2-3 times
faster than Oracle in most cases.
• Indexes in MongoDB did not seem to have any
significant effect on performance.
• When running queries in parallel MongoDB per-
forms better for Test Case 1 and Test Case 2. Or-
acle kept the same query/s ratio (which resulted
in exponential growth of test duration, depending
on the number of threads). Query/s ratio of Mon-
goDB was increasing significantly up to 8 threads
(and it kept the ratio for 16 and 32 threads)
• MongoDB had problems with Test Case 3 under
heavy load (16 and 32 threads).
5.2 Flexibility
Due to its query system and document nature, Mon-
goDB is suitable for searching experimental meta-
data. However, it does not provide an ultimate solu-
tion for all experimental metadata as there are pieces
of information for which relational integrity is re-
quired (typically a person own several experiment or
one scenario can be used in more experiments (see
Figure 1)). MongoDB does support foreign keys, yet
their usage would result in multiple queries (as it does
not support joins).
There is several options, each with own difficulties
to overcome:
1. Keep all data in a relational database - It ensures
an easy way to maintain data integrity, but it does
not provide a flexible data model.
2. Transfer all data to MongoDB - It ensures a flexi-
ble data model but it brings many difficulties. Ei-
ther it contains duplicate data or references due
to nonexistent foreign keys. Some application
changes are required. Although, it brings fast read
queries, a need for multiple queries (in case of ref-
erences usage) to retrieve all related data to an in-
dividual record occurs.
3. Parallel use of both database engines
(a) in one application - It requires to keep data
which require relational integrity in Oracle and
to keep experiment-specific meta-data in Mon-
goDB.
(b) separate data warehouse - architecture taken
from the StorageBIT (Carreiras et al., 2013)
proposal requires implementation a separate
experiment data storage on top of MongoDB,
and access it from the Portal via public API.
6 DISCUSSION
From Section 5.1 we can observe that Oracle per-
formed better only in Test Case 3. One of the rea-
sons might be that while Oracle can get the age value
from a date, MongoDB has to compare dates directly.
MongoDB gave stable results for all iterations. Ora-
cle experienced significant performance jumps during
iterations, for unknown reason (as a result the average
query/s ratio contains rather high statistical error).
Option 2 from Section 5.2 seems to be the least
suitable solution for the project such as the EEG/ERP
Portal. Even though the NoSQL database provides
good facility for storing electrophysiology experi-
ments, it is not the best choice for the current version
of the EEG/ERP Portal. Any attempt to use solely
MongoDB as the EEG/ERP Portal’s data storage, due
different nature of NoSQL databases, would probably
result in large changes in the application.
Option 1 is the safest one, while option 3 is worth
further investigation as it might result in a solution
which would be able to combine strengths of both
concepts. Option 3a is most difficult to implement
(probably most complicated changes to the appli-
cation requires answer questions how to query ef-
ficiently and how to handle transactions?). Option
3b would require changes in the Portal’s application
structure. In addition, it brings similar integrity issues
as with 3a. The recommended approach uses combi-
nation of Hibernate (Bauer and King, 2006) for Or-
acle and Spring Data (Pollack et al., 2012) for Mon-
goDB. A layer implemented between the application
and persistence provides a unified access to data in a
relational database and data in a NoSQL database.
7 CONCLUSIONS
Lot of difficulties were identified when electrophys-
iological data were stored and maintained. The aim
of current research initiatives is to design robust sys-
tems for storing large collections of data sets. We de-
veloped the EEG/ERP Portal. The robustness of this
MongoDBforElectrophysiologyExperiments
425

Table 1: Performance results. It compares Oracle (the left part of table) with MongoDB (the right part of table) for all three
test cases defined in Section 5.1. The upper part of the table shows results when indexes were disabled, the bottom part when
indexes were enabled.
Oracle MongoDB
Without indexes
Test Case 1
Processes 1 2 4 8 16 32 1 2 4 8 16 32
avg(s) 15,749 29,373 59,722 131,443 244,95 320,111 6,899 7,612 8,24 13,42 25,863 52,082
op 100 200 400 800 1600 3200 100 200 400 800 1600 3200
op/s 6,35 6,809 6,698 6,086 6,532 9,997 14,495 26,273 48,544 59,615 61,865 61,442
avg 7,079 45,372
Test Case 2
avg(s) 17,984 32,422 57,652 128,311 220,652 131,534 9,891 10,121 10,557 18,618 36,827 74,206
op 100 200 400 800 1600 3200 100 200 400 800 1600 3200
op/s 5,561 6,169 6,938 6,235 7,251 24,328 10,111 19,761 37,888 42,969 43,446 43,123
avg 9,414 32,883
Test Case 3
avg(s) 72,404 143,456 286,598 387,175 323,436 514,247 23,946 45,673 80,695 163,815 373,29 753,373
op 100 200 400 800 1600 3200 100 200 400 800 1600 3200
op/s 1,381 1,394 1,396 2,066 4,947 6,223 4,176 4,379 4,957 4,884 4,286 4,248
avg 2,901 4,488
With indexes
Test Case 1
Processes 1 2 4 8 16 32 1 2 4 8 16 32
s 13,996 26,683 48,646 84,48 164,857 304,819 6,446 7,21 7,767 12,604 24,271 48,852
op 100 200 400 800 1600 3200 100 200 400 800 1600 3200
op/s 7,145 7,495 8,223 9,47 9,705 10,498 15,513 27,74 51,503 63,474 65,921 65,504
avg 8,756 48,276
Test Case 2
s 16,45 30,518 57,537 118,879 150,591 140,543 9,336 9,68 10,142 17,374 34,471 69,582
op 100 200 400 800 1600 3200 100 200 400 800 1600 3200
op/s 6,079 6,553 6,952 6,73 10,625 22,769 10,711 20,66 39,439 46,045 46,416 45,989
avg 9,951 34,877
Test Case 3
s 64,773 128,272 255,519 379,731 338,034 500,823 21,885 44,654 81,854 167,622 399,704 828,011
op 100 200 400 800 1600 3200 100 200 400 800 1600 3200
op/s 1,544 1,559 1,565 2,107 4,733 6,389 4,569 4,479 4,887 4,773 4,003 3,865
avg 2,983 4,429
system is ensured using Oracle database on backend.
Although this solution works quite satisfactorily, a us-
age of relational database makes difficult to store het-
erogeneous experimental data. This disadvantage is
solved using document-based databases. We tested
MongoDB and investigate possibilities to use it in-
stead of Oracle. Despite the advantages that this so-
lution brings, a lot of difficulties remain. The most
significant is the loss of data integrity together with a
need to duplicate data items. The second one is an ab-
sence of a unified API to access data (some solution
similar to Hibernate that ensures object-relational-
mapping for Java programs). Our future work in-
cludes two following goals: The data where its per-
sistence is strictly required will be exactly defined.
Next, a unified API that enables parallel data access
in a fixed schema database together with data access
in a schema free database will be provided.
ACKNOWLEDGEMENTS
This work was supported by the European Regional
Development Fund (ERDF), Project ”NTIS - New
Technologies for Information Society”, European
Centre of Excellence, CZ.1.05/1.1.00/02.0090.
REFERENCES
Bauer, C. and King, G. (2006). Java Persistence with Hi-
bernate. Manning, revised. edition.
Carreiras, C., Silva, H., Loureno, A., and Fred, A. L. N.
(2013). Storagebit - a metadata-aware, extensible, se-
mantic and hierarchical database for biosignals. In
Stacey, D., Sol-Casals, J., Fred, A. L. N., and Gamboa,
H., editors, HEALTHINF, pages 65–74. SciTePress.
Grewe, J., Wachtler, T., and Benda, J. (2011). A bottom-up
HEALTHINF2014-InternationalConferenceonHealthInformatics
426

approach to data annotation in neurophysiology. Fron-
tiers in Neuroinformatics, 5(16).
Group, I. W. (2013). Incf dataspace.
Jezek, P. and Moucek, R. (2012). SYSTEM FOR
EEG/ERP DATA AND METADATA STORAGE
AND MANAGEMENT. NEURAL NETWORK
WORLD, 22(3):277–290.
Jezek, P., Stebet
´
ak, J., Bruha, P., and Moucek, R. (2013).
Model of software and hardware infrastructure for
electrophysiology. In Stacey, D., Sol
´
e-Casals, J., Fred,
A. L. N., and Gamboa, H., editors, HEALTHINF,
pages 352–356. SciTePress.
Leavitt, N. (2010). Will nosql databases live up to their
promise? Computer, 43(2):12–14.
Pollack, M., Gierke, O., Risberg, T., Brisbin, J., and
Hunger, M. (2012). Spring Data. O’Reilly Media.
Rautenberg, P., Sobolev, A., Herz, A., and Wachtler, T.
(2011). A database system for electrophysiological
data. In Hameurlain, A., Kng, J., Wagner, R., Bhm,
C., Eder, J., and Plant, C., editors, Transactions on
Large-Scale Data- and Knowledge-Centered Systems
IV, volume 6990 of Lecture Notes in Computer Sci-
ence, pages 1–14. Springer Berlin Heidelberg.
The HDF Group (2000-2010). Hierarchical data format ver-
sion 5.
van Pelt, J. and van Horn, J. (2007). 1st incf workshop
on sustainability of neuroscience databases. Workshop
report. Stockholm.
MongoDBforElectrophysiologyExperiments
427
