
BRIDG-based Trial Metadata Repository
Need for Standardized Machine Interpretable Trial Descriptions
J. van Leeuwen
1
, A. Bucur
1
, B. Claerhout
2
, K. De Schepper
2
, D. Perez-Rey
3
and R. Alonso-Calvo
3
1
HIM department, Philips Electronics BV, High Tech Campus 34, Eindhoven, the Netherlands
2
Custodix NV, Kortrijksesteenweg 214b3, Sint-Martens-Latem, Belgium
3
Departamento de Inteligencia Artificial, Universidad Politécnica de Madrid, Boadilla del Monte, Madrid, Spain
Keywords: Clinical Trials, Trial Metadata, Trial Metadata Repository, Semantic Interoperability, Trial Eligibility.
Abstract: Making information about clinical trials accessible in a machine interpretable way could aid applications
both in clinical care and clinical research, such as patient screening, trial recruitment, trial meta-analysis,
trial duplication detection and clinical decision support. We present our standards-based trial metadata
repository that captures structured trial information and application-specific formalisms and execution logic
supporting a range of relevant applications, with a focus on interoperability and machine interpretability to
enable more efficient support for clinical research and faster knowledge transfer into care, We further
exemplify the use of the Trial Metadata Repository for a patient screening application for clinical trials.
Additionally, the mechanisms are described to manage the information model of the repository when the
scope is enlarged to additional contexts.
1 INTRODUCTION
From academic medical research centres to
community hospitals and other stakeholders, the
healthcare industry continues to improve its
capabilities for electronic data capture. Ideally,
clinical care and clinical research would live
symbiotically together, resulting in optimal patient
care –based on the latest validated research
findings– and efficient increase of clinical
knowledge (aided by the accessibility of clinical care
information). Currently however, there is a large
separation between clinical care and clinical
research with information typically silo-ed in the
respective contexts.
The execution of clinical trials is an important
vehicle used in clinical research to progress clinical
knowledge. In addition, treatment in a clinical trial is
often a cancer patient’s best option (Edwards, 1998).
The National Comprehensive Cancer Network
advises that the best management of any cancer
patient is in a clinical trial and encourages
participation in clinical trials (NCCN, 2010).
Unfortunately, comprehensive information about
clinical trials is not easily accessible at the point of
care. Typically, the clinical trial protocol is only
accessible in a non-machine-interpretable form (such
as paper or pdf file). The clinical trial protocol
describes amongst others the purpose of the trial, the
clinical rationale, eligibility criteria, and the
schedule and details of the tests, procedures, and/or
medication.
In addition, information about the results of
clinical trials is inefficiently transferred back to
clinical care. Again, information is accessible in a
non-machine-interpretable form (typically in the
form of literature, papers or guidelines) which is
time-consuming to process for the clinical users and
cannot be used for automatic processing in relevant
applications.
As the information is in a non-machine-
interpretable form, it is also not possible to aid the
clinician by targeting the information to the patient
case at hand in a clinical decision support
application.
Capturing the information about clinical trials in
a machine interpretable way could aid applications
both in clinical care and clinical research. For
instance, it could aid a clinician to find a suitable
trial for a patient (patient screening), it could aid a
clinical researcher and a pharmaceutical company to
efficiently recruit participants (trial recruitment), it
453
van Leeuwen J., Bucur A., Claerhout B., De Schepper K., Perez-Rey D. and Alonso-Calvo R..
BRIDG-based Trial Metadata Repository - Need for Standardized Machine Interpretable Trial Descriptions.
DOI: 10.5220/0004910304530458
In Proceedings of the International Conference on Health Informatics (HEALTHINF-2014), pages 453-458
ISBN: 978-989-758-010-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

could increase the medical knowledge by allowing
more efficient data analysis across trials (meta-
analysis), it could prevent duplicating the execution
of trials (duplicate detection) and it could aid a
clinician in finding relevant treatment options for a
patient (clinical decision support). Therefore, an
effective solution is required to represent structure
and store this information. We name the repository
storing this information the trial metadata repository.
By convention we call this information the trial
metadata to differentiate from the term “trial data”
which typically refers to the patient data collected
for a clinical trial.
1.1 State of Practice
The realization that information about clinical trials
should be publically available is nowadays common
ground. The World Health Organization (WHO)
publishes the WHO Trial Registration Data Set
(International Clinical Trials Registry Platform,
2013) which specifies the minimum amount of trial
information that must appear in a trials registry. The
WHO site also contains a collection of links to trial
registries (which are typically organized on a
geographical level).
In addition, various countries have made
legislation enforcing companies to publish clinical
trial information.
In the current state of practice, many trials
publish information on clinicaltrials.gov (i.e.
disease, intervention, eligibility criteria, etc.).
Unfortunately, these initiatives are focused on a
textual distribution of the information. We argue that
the information should be made accessible in a
machine interpretable way, allowing for
contextualization of the information given and
enabling a wide range of applications that rely on
access to structured trial information, such as
clinical decision support, trial recruitment, meta-
analysis of trial results, duplicate trial design
detection, etc.
Existing initiatives like linkedct.org (which aims
at publishing an open Semantic Web data source for
clinical trials data) are of limited use as the
information is post-processed from clinicaltrials.gov
and is rather course grained. To illustrate this, the
following criteria text excerpt has been retrieved
from linkedct.org, which is available as blob only
(i.e. not structured):
“DISEASE CHARACTERISTICS: - Histologically
proven metastatic renal cell carcinoma not
amenable to complete surgical resection and
progressive despite immunotherapy -
Bidimensionally evaluable clinically or
radiographically - HLA 6/6 or 5/6 matched family
donor available - No CNS metastases PATIENT
CHARACTERISTICS: Age: - 18 to 80 Performance
status: - ECOG 0 or 1 Life expectancy: - At least 3
months Hematopoietic: - Not specified Hepatic: -
Bilirubin no greater than 4 mg/dL - Transaminases
no greater than 3 times upper limit of normal Renal:
- Creatinine no greater than 2.5 mg/dL - No
malignancy-associated hypercalcemia (< 2.5
mmol/L) Cardiovascular: - Left ventricular ejection
fraction greater than 40% Pulmonary: - DLCO
greater than 65% of predicted Other: - Not pregnant
- HIV negative - No major organ dysfunction that
would preclude transplantation - No other
malignancies except basal cell or squamous cell skin
cancer - No psychiatric disorder or mental
deficiency that would preclude study participation
PRIOR CONCURRENT THERAPY: Biologic
therapy - See Disease Characteristics Chemotherapy
- Not specified Endocrine therapy - Not specified
Radiotherapy - Not specified Surgery - Not specified
Other - At least 1 month since prior treatment for
renal cell carcinoma.” (Hassanzadeh, 2013). This
unfortunately does not allow for contextualization or
processing.
At the same time, the Biomedical Research
Integrated Domain Group (BRIDG) Model
initiative (Biomedical Research Integrated Domain
Group Model, 2013) is gaining traction. The BRIDG
model is a domain analysis model which aims to
provide a shared view of the dynamic and static
semantics for the domain of protocol-driven research
and its associated regulatory artifacts. The BRIDG
model is a collaborative effort spanning important
and relevant standardization bodies like the Clinical
Data Interchange Standards Consortium (CDISC),
the HL7 Regulated Clinical Research Information
Management Technical Committee (RCRIM) Work
Group, the US National Cancer Institute (NCI), and
the US Food and Drug Administration (FDA). This
collection of stakeholders ensures a wide variety of
viewpoints on the model, which increases the
potential for stability of the model. In addition the
BRIDG model has the promise of easing future
interoperability as the various standardization bodies
are defining their new standards based in the BRIDG
model. As the BRIDG model is a domain analysis
model (and a conceptual model for clinical
research), it cannot be used “as is” to implement a
physical design or to generate code. Rather it can be
leveraged to further build out detailed logical models
and physical designs.
BRIDG currently spans the following specialized
HEALTHINF2014-InternationalConferenceonHealthInformatics
454
subdomains: Protocol representation focusses on
planning and design of a research protocol, Study
conduct focusses on the execution of a research
protocol (study conduct and results from the study
activities), Statistical analysis describes the planning
and performance of the statistical analysis of data
collected during clinical trial research and their
relationships, Adverse event focusses on all safety
related activities, e.g. detection, evaluation, follow-
up and reporting, regulatory focusses on
submissions to regulatory authorities.
2 THE TRIAL METADATA
REPOSITORY
In order to support the efficient dissemination of
clinical trial information, interoperability and
machine-interpretability of content should be
important features of a trial metadata repository.
In the context of the INTEGRATE project
(www.fp7-integrate.eu) – a collaboration project
aiming to develop innovative infrastructures to
enable data and knowledge sharing and to foster
large-scale collaboration in biomedical research –
the need arose for a trial metadata repository. The
developed trial metadata repository leverages the
BRIDG domain analysis model for its information
model to facilitate interoperability.
The trial metadata repository uses the BRIDG
domain analysis model by subsetting the model
(selecting the concepts and relations necessary for
the use cases) and subsequently extending the set
with application specific concepts.
The information model for the trial metadata
repository is expressed in the Unified Modeling
Language (UML) (www.uml.org) – a modeling
language that includes a set of graphical notation
techniques to create visual models of object oriented
software-intensive systems (Unified Modeling
Language, 2013) – as is the BRIDG model.
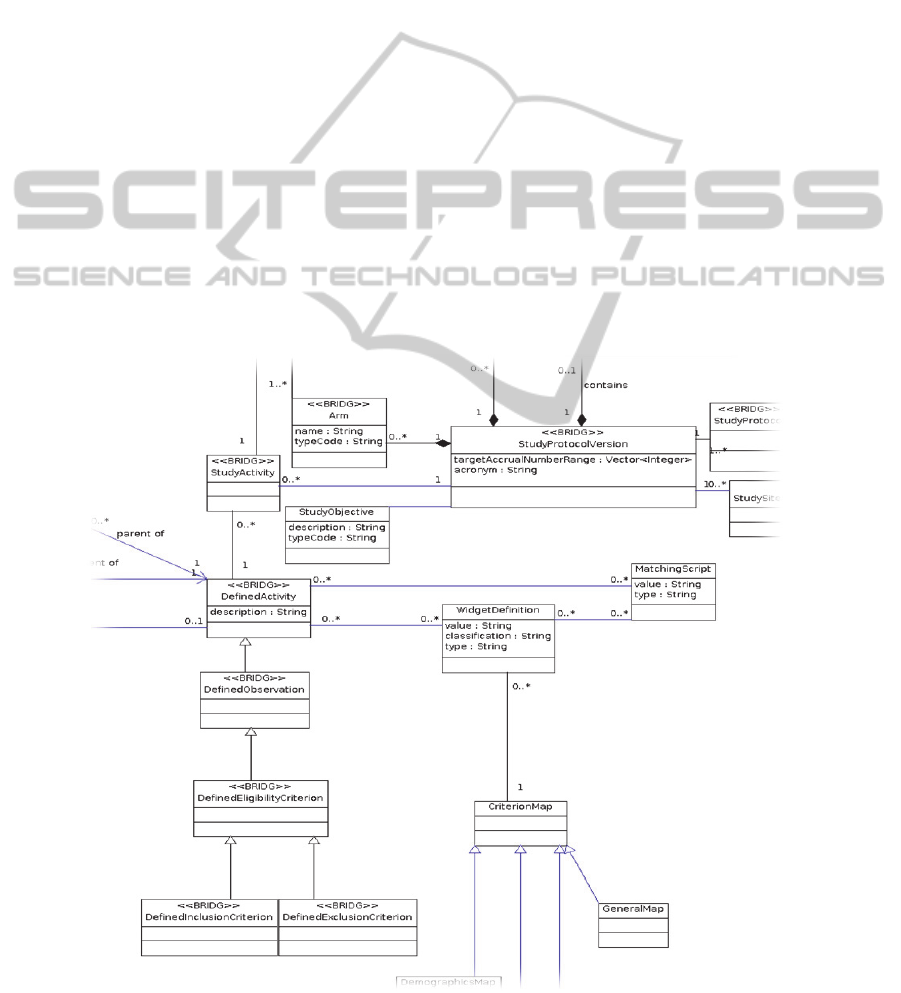
Figure 1 shows an excerpt of the information
model, depicting a version of a study protocol and
its relations to the inclusion and exclusion criteria.
The model is extended with application specific
information where our use cases require trial
metadata (currently) not covered by the BRIDG
model.
Figure 1: Information model excerpt.
BRIDG-basedTrialMetadataRepository-NeedforStandardizedMachineInterpretableTrialDescriptions
455

Figure 2: Script example evaluating the eligibility of a patient requiring the GPT lab value to be less than 1.5 * max.
The trial metadata repository has an administrative
interface enabling users to inspect, input and update
trial descriptions in the trial metadata repository.
This enables to efficiently populate the repository
with new trials, implement changes (e.g. for
approved trial amendments), extend the solution
with new formalisms and deploy the trial metadata
repository for new applications.
2.1 Trial Eligibility Evaluation
A current application for which the trial metadata
repository is essential addresses the use case of a
clinician assessing the eligibility of a patient for
enrolment into a particular clinical trial. In order to
enrol into a clinical trial, the patient must meet the
eligibility criteria of the trial (covering specific
criteria like cancer type, previous treatments, health
status, etc.). The eligibility criteria and their
verification are typically not integrated into the
clinical information systems, but usually the trial
description (with the eligibility criteria) is
distributed as a read-only document (be it
electronically or in print). To verify clinical trial
eligibility, the clinician has to browse through the
clinical information systems in order to find the
required patient information so eligibility status can
be assessed (which can be a time consuming
activity).
In order to provide maximum benefit, the trial
metadata repository should not only provide easy
access to the trial information, but it should also
provide a means to easily connect the information
with patient data. For the trial eligibility use case,
the trial metadata repository heavily leverages
classes from the Protocol Representation of BRIDG
to capture information about clinical trials such as
name and description of the trial, recruitment status,
inclusion and exclusion criteria, current patient
accrual, target patient accrual, due date, etc.
In addition, the information model has been
extended with the ability to associate statements in
different formalisms with each criterion. The
required formalism might differ depending on how
the patient data is stored (syntax and semantics) and
how the patient data is accessible (e.g. using
def query="""SELECT DISTINCT ?obs_id ?value ?max
WHERE {
?instParti a hl7rim:participation;
hl7rim:participation_entityId "$patientID";
hl7rim:participation_act ?instAct.
?instAct hl7rim:act_observation ?instObs.
?instObs hl7rim:observation_code "34608000";
hl7rim:observation_id ?obs_id;
hl7rim:observation_valueST ?value;
hl7rim:observation_refRangeMax ?max;
}""";
Map<String,String> values = semanticLayerWSClientImpl.executeQuery(target,
query);
QueryResult[] result = new QueryResult[1];
result[0] = new QueryResult();
if(values == null){
result[0].setResult(MatchResult.UNDETERMINED);
}else{
if (values.get("value")==null || values.get("max")==null) {
result[0].setResult(MatchResult.UNDETERMINED);
return result; }
String id = values.get("obs_id");
result[0].setResult(MatchResult.NONMATCH);
if (new Float(values.get("value")) < (1.5* new Float(values.get("max"))) ){
result[0].setResult(MatchResult.MATCH);}
Evidence evidence = new Evidence();
evidence.setEvidenceId(id);
result[0].setEvidence(evidence);
};
return result;
HEALTHINF2014-InternationalConferenceonHealthInformatics
456
webservices/odbc/sparql/etc.). For the trial eligibility
application, the trial metadata repository stores
executable logic (in the example below a groovy
script). In order to bridge the gap between trial
information and patient data, standard
ontologies/terminologies are used in the formalisms
to achieve shared semantics.
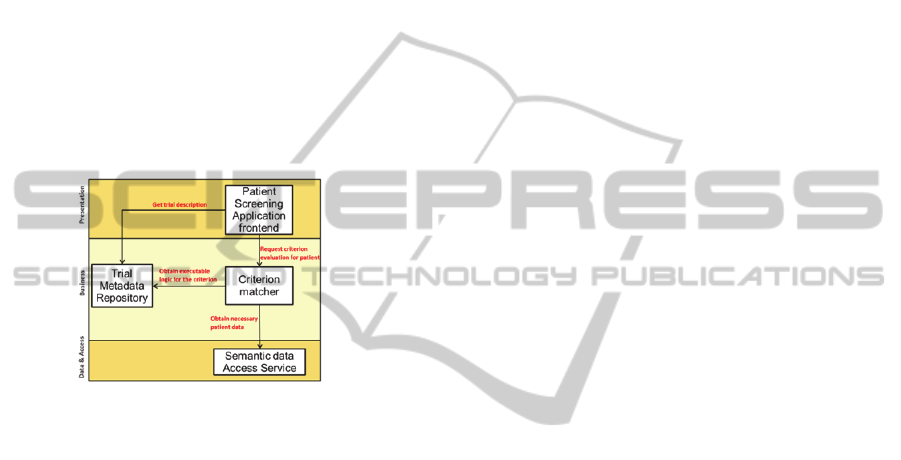
The INTEGRATE trial eligibility application is
composed of the following components: The
application front-end: the user interface for the
clinical care giver; the criterion matcher: retrieves
the executable logic for a criterion and executes it to
assess eligibility, the trial metadata repository:
contains the trial information and executable logic
for the eligibility criteria, and the semantic data
access service: (semantically enabled) data access
services for patient data.
Figure 3: architecture of the trial eligibility application.
The development of the trial metadata repository
is based on a model driven approach. This choice
has been made as it is expected that the information
model will be regularly extended according the
needs of additional use cases. Technically, a series
of code generators have been created that transform
the UML description of the information model into
the actual trial metadata repository.
The code generators take as input an xml
serialization (xmi) of the UML model and
subsequently:
create the underlying database
expose the content via webservices
create an administrative web interface
create the documentation of the information
model.
The components comprising the trial eligibility
demonstrator are loosely coupled and the
architecture is a Service Oriented Architecture. The
deployment of the components is flexible and the
trial metadata repository can be deployed in various
context – e.g. in a hospital context, enterprise
context or across enterprises.
In the trial eligibility application, executable
logic is used as formalism. Each criterion is
associated with a groovy script which will be
executed by the criterion matcher. The script
typically contains queries to retrieve the necessary
patient data and subsequently uses that data to
evaluate the criterion. As example a script (Figure 2)
is described that evaluates a laboratory value (the lab
value for GPT (glutamate pyruvate transaminase, an
enzyme measurement associated with liver
functioning) should be lower than 1.5 times the
upper value of the normal range of the laboratory).
The script contains a SPARQL query to retrieve the
relevant patient data. Shared semantics are used in
this demonstrator, binding ah HL7v3 RIM based
information model with an core dataset consisting of
SNOMED, MedDRA and LOINC, and in the
example an observation with SNOMED code
"34608000” (the GPT lab value) is retrieved. The
criterion matcher inserts the correct patient id and
retrieves the observation id (obs_id), the value
(value) and the upper value of the normal value of
range of the lab (max). Subsequently, the scripts
evaluates the criterion (by evaluating whether value
< i.5* max.) Finally, it returns whether there is a
match, a non-match, or that the criterion could not
be evaluated.
2.2 Current and Future Work
We plan to further extend the trial metadata
repository for other use cases, to capture information
about the structure of the clinical trial (a shared
standard representation of the components of a trial)
and to capture the results of trials.
As discussed in (Speedie et al., 2008): “A
standard representation of Clinical trial information
is necessary to clearly and accurately communicate
the structure of a trial for uniform implementation at
multiple sites. One of the challenges in such
multisite trials is consistent implementation, when
numerous individuals at the different sites are
charged with executing the trial. Inconsistencies can
arise from different understandings of the protocol’s
elements. Consistency is supported by a common
understanding of the relevant aspects of the trial.
Aside from needing a standard representation of
clinical trial information to help run a trial, such a
representation is essential for combining results
from multiple heterogeneous clinical trials in a
meta-analysis, where small differences in trial
design and outcome measures may lead to
inaccuracy in the overall effect estimate. The ability
to determine which elements of two or more trials
BRIDG-basedTrialMetadataRepository-NeedforStandardizedMachineInterpretableTrialDescriptions
457

are similar and which are different is critical to
detecting such differences. Without a standard
method of representing the components of a trial, it
is necessary to depend entirely on the interpretations
of readers regarding the comparability of trial
elements. There is an overlapping and equally
important issue of the standard representation and
reporting of clinical data for the purposes of
comparing the results of multiple clinical studies.
Essential to the task of conducting a systematic
review of clinical trials is the need to objectively
evaluate the quality of the trials. For this task, it is
important to be able to understand the design
elements of a given trial and be able to compare it
with others of known quality. These comparisons
require identification and description of trial
components such as treatment allocation strategies,
in clear and unambiguous terms, to make valid
judgments about the overall trial quality. The lack of
a standard representation of trial design features
impedes this process by making it more difficult to
locate and characterize the important elements of a
trial that are used in critical appraisals of trial
evidence. A standard, computable representation
would improve the ability to evaluate the quality of
clinical trials and provide a basis for doing so in an
automated fashion”.
These additions could increase the medical
knowledge by allowing more efficient data analysis
across trials (meta-analysis), it could prevent
execution of similar trials (duplicate detection) and it
could aid a clinician in finding relevant treatment
options for a patient (clinical decision support).
With the addition of more and more types of
information to the information model, it becomes
important to manage data capture in a coherent way.
Information should be sound and complete. In order
to aid the user in capturing sound and complete
information, views will be provided on the
information. Views will be pertinent to specific
application areas - like trial recruitment (with a
focus on eligibility criteria) or trial (meta-)analysis
(with a focus on the components of a trial design).
These views will be captured in the UML model
by the use of tagged values (allowing to add
additional information to UML elements),
leveraging a dedicated UML profile.
3 CONCLUSIONS
The information about clinical trials that is currently
locked away in non-machine-interpretable form
(typically .pdf or paper) can deliver a lot of value to
a wide variety of application in the clinical care and
clinical research domains. We have presented our
ongoing work to unlock this information. For this,
we have devised an information model leveraging
the BRIDG model (to ensure future interoperability)
and implemented a trial metadata repository that
stores in a structured, semantics-aware way relevant
trial information. This information model has been
extended to allow storing different application
specific formalisms and execution logic, for instance
to describe machine-interpretable eligibility criteria.
These additional elements have the role to support a
variety of applications that need access to trial
information.
Finally, we describe how the information model
can be extended for different clinical trial contexts
while ensuring maintainability.
In our future work we will extend the current
solution to support a wide range of applications in
clinical research and clinical care.
ACKNOWLEDGEMENTS
This work has been partially funded by the European
Commission through the INTEGRATE project
(FP7-ICT-2009-6-270253).
REFERENCES
Hassanzadeh, O. (2013). Retrieved 11 12, 2013, from The
Linked Clinical Trials (LinkedCT) project:
http://data.linkedct.org/resource/eligibility/52af484a1a
ea77150c3a9f454226b302/
(2013). Retrieved 11 12, 2013, from Biomedical Research
Integrated Domain Group Model:
http://bridgmodel.nci.nih.gov/
International Clinical Trials Registry Platform. (2013).
Retrieved 11 12, 2013, from WHO:
http://www.who.int/ictrp/network/trds/en/index.html
Unified Modeling Language. (2013). Retrieved 11 12,
2013, from Wikipedia: http://en.wikipedia.org/wiki/
Unified_Modeling_Language
Edwards, M. (1998). Access to quality care: Concensus
statement of the american federation of clinical
oncologic societies. Annals of Surgical Oncology,
657-659.
NCCN. (2010). NCCN Clinical Practie Guidelines in
Oncology - Breast Cancer.
Speedie, S., Taweel, A., Sim, I., Arvanitis, T., Delaney,
B., & Peterson, K. (2008). The Primary Care Research
Object Model (PCROM): A Computable Information
Model for Practice-based Primary Care Research. J
Am Med Inform Assoc., 661–670.
HEALTHINF2014-InternationalConferenceonHealthInformatics
458
