
Novel Feature Selection Methods for High Dimensional Data
Ver´onica Bol´on-Canedo, Noelia S´anchez-Maro˜no and Amparo Alonso-Betanzos
Department of Computer Science, University of A Coru˜na, Campus de Elvi˜na s/n, A Coru˜na 15071, Spain
1 INTRODUCTION AND STATE
OF THE ART
In the past 20 years, the dimensionality of the datasets
involved in data mining has increased dramatically,
as can be seen in (Zhao and Liu, 2011). This fact is
reflected if one analyzes the dimensionality (samples
× features) of the datasets posted in the UC Irvine
Machine Learning Repository (Frank and Asuncion,
2010). In the 1980s, the maximal dimensionality of
the data was about 100; then in the 1990s, this number
increased to more than 1500; and finally in the 2000s,
it further increased to about 3 million. The prolifera-
tion of this type of datasets with very high (> 10000)
dimensionality has brought unprecedented challenges
to machine learning researchers. Learning algorithms
can degenerate their performance due to overfitting,
learned models decrease their interpretability as they
are more complex, and finally speed and efficiency of
the algorithms decline in accordance with size.
Machine learning can take advantage of feature
selection methods to be able to reduce the dimension-
ality of a given problem. Feature selection (FS) is the
process of detecting the relevant features and discard-
ing the irrelevant and redundant ones, with the goal
of obtaining a small subset of features that describes
properly the given problem with a minimum degra-
dation or even improvement in performance (Guyon
et al., 2006). Feature selection, as it is an important
activity in data preprocessing, has been an active re-
search area in the last decade, finding success in many
different real world applications, especially those re-
lated with classification problems.
There are several situations that can hinder the
process of feature selection, such as the presence of
irrelevant and redundant features, noise in the data or
interaction between attributes. In the presence of hun-
dreds or thousands of features, such as DNA microar-
ray analysis, researchers notice (Yu and Liu, 2004)
that is common that a large number of features is not
informative because they are either irrelevant or re-
dundant with respect to the class concept. Moreover,
when the number of features is high but the number of
samples is small, machine learning gets particularly
difficult, since the search space will be sparsely popu-
lated and the model will not be able to distinguish cor-
rectly the relevant data and the noise (Provost, 2000).
Feature selection methods usually come in three
flavors: filter, wrapper, and embedded methods
(Guyon et al., 2006). The filter model relies on the
general characteristics of training data and carries out
the feature selection process as a pre-processing step
with independence of the induction algorithm. On the
contrary, wrappers involve optimizing a predictor as
a part of the selection process. Halfway these two
models one can find embedded methods, which per-
form feature selection in the process of training and
are usually specific to given learning machines. By
having some interaction with the predictor, wrapper
and embedded methods tend to obtain higher predic-
tion accuracy than filters, at the cost of a higher com-
putational cost.
There exist numerous papers and books proving
the benefits of the feature selection process (Guyon
et al., 2006; Dash and Liu, 1997; Kohavi and John,
1997; Zhao and Liu, 2011). However, most re-
searchers agree that there is not a so-called “best
method” and their efforts are focused on finding a
good method for a specific problem setting. There-
fore, new feature selection methods are constantly
emerging using different strategies: a) combining sev-
eral feature selection methods, which could be done
by using algorithms from the same approach, such as
two filters (Zhang et al., 2008), or coordinating algo-
rithms from two different approaches, usually filters
and wrappers (Peng et al., 2010); b) combining fea-
ture selection approaches with other techniques, such
as feature extraction (Vainer et al., 2011) or tree en-
sembles (Tuv et al., 2009); c) reinterpreting existing
algorithms (Sun and Li, 2006), sometimes to adapt
them to specific problems (Sun et al., 2008); d) cre-
ating new methods to deal with still unresolved situ-
ations (Chidlovskii and Lecerf, 2008; Loscalzo et al.,
2009) and e) using an ensemble of feature selection
techniques to ensure a better behavior (Saeys et al.,
2008).
3
Bolón-Canedo V., Sánchez-Maroño N. and Alonso-Betanzos A..
Novel Feature Selection Methods for High Dimensional Data.
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)
2 RESEARCH PROBLEM
As mentioned in the introduction, in the last years
the dimensionality of datasets involved in data mining
applications has increased steadily. This large-scale
data carries new opportunities and challenges to com-
puter scientists, giving the opportunity for discover-
ing subtle population patterns and heterogeneities that
were not possible with small-scale data. However,
the massive sample size and high dimensionality of
data introduce new computational challenges. The-
oretically, having more data should give more dis-
criminating power. However, the nature of high di-
mensionality of data can cause the so-called problem
of curse of dimensionality or Hughes effect (Hughes,
1968). This phenomenon occurs when the model has
to be learned from a finite number of data samples
in a high-dimensional feature space with each feature
having a number of possible values, and so an enor-
mous amount of training data are required to ensure
that there are several samples with each combination
of values. The Hughes effect is therefore known as
the situation where with a fixed number of training
samples, the predictive power of the learner reduces
as the feature dimensionality increases. In this situa-
tion, feature selection plays a crucial role.
There exists a vast body of feature selection meth-
ods in the literature, including filters based on distinct
metrics (e.g. entropy, probability distributions or in-
formation theory) and embedded and wrappers meth-
ods using different induction algorithms. The prolif-
eration of feature selection algorithms, however, has
not brought about a general methodology that allows
for intelligent selection from existing algorithms. In
order to make a correct choice, a user not only needs
to know the domain well, but also is expected to un-
derstand technical details of available algorithms (Liu
and Yu, 2005). On top of this, most algorithms were
developed when dataset sizes were much smaller, but
nowadays distinct compromises are required for the
case of small-scale and large-scale (big data) learning
problems. Small-scale learning problems are subject
to the usual approximation-estimation trade-off. In
the case of large-scale learning problems, the trade-
off is more complex because it involves not only the
accuracy of the selection but also other aspects, such
as stability (i.e. the sensitivity of the results to training
set variations) or scalability.
The objective of this research is two-fold. First,
an analysis of classical feature selection is performed,
evaluating the adequacy of different methods in dif-
ferent situations. Moreover, the benefits of feature se-
lection have been studied in different data settings:
a) datasets with a number of samples much higher
than the number of features; b) datasets with a number
of features much higher than the number of samples;
and c) datasets with a high number of features and
samples. After studying the feature selection domain,
the second part of the research is devoted to develop
novel feature selection to be applied to high dimen-
sional datasets.
3 OUTLINE OF OBJECTIVES
As mentioned above, the main goals of the thesis are
to analyze in detail the feature selection domain and
then, to develop novel feature selection methods for
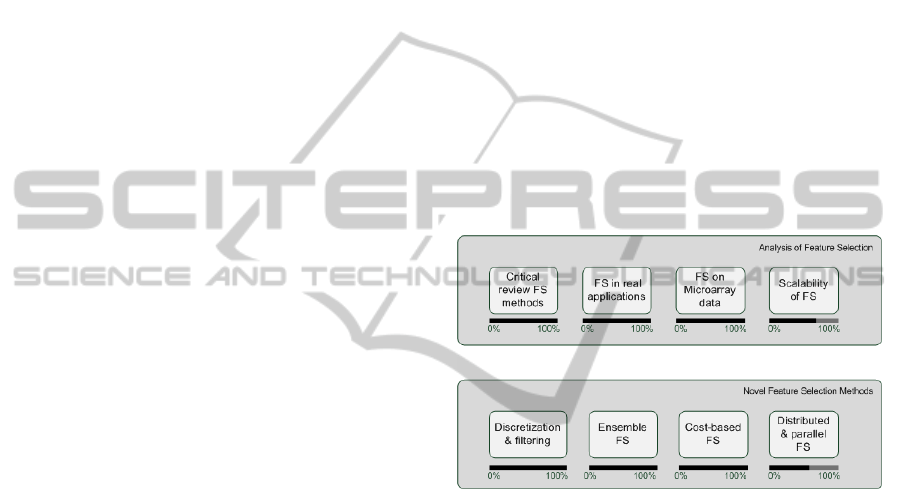
high-dimensional data. An outline of this can be seen
in Figure 1, where each objective is divided in several
subobjectives, which will be following described in
detail. Notice that the diagram also includes informa-
tion about the level of completion of each task.
Figure 1: Outline of objectives.
1. Analysis of classical feature selection and appli-
cation to real problems:
(a) Critical review of the most popular feature se-
lection methods in the literature by checking
their performance in an artificial controlled ex-
perimental scenario. In this manner, the abil-
ity of the algorithms to select the relevant fea-
tures and to discard the irrelevant ones without
permitting noise or redundancy to obstruct this
process is evaluated.
(b) Application of classic feature selection to real
problems in order to check their adequacy.
Specifically, testing the effectiveness of feature
selection in two problems found in the medical
domain: tear film lipid layer classification and
K-complex identification in sleep apnea.
(c) Analysis of the behavior of feature selection
in a very challenging field: DNA microarray
classification. DNA microarray data is a hard
challenge for machine learning researchers due
to the high number of features (around 10 000)
ICAART2014-DoctoralConsortium
4

but small sample size (typically one hundred or
less). For this purpose, it is necessary to re-
view the most up-to-date algorithms developed
ad-hoc for this type of data, as well as studying
their particularities.
(d) Analysis of the scalability of existing feature
selection methods. With the advent of high-
dimensionality, machine learning researchers
are not focused only in the accuracy of the se-
lection, but also in the scalability of the solu-
tion. Therefore, this issue must be addressed,
covering the three situations described in Sec-
tion 2: high number of samples, high number of
features, high number of samples and features.
2. Development of novel feature selection methods
for high-dimensional data
(a) Development of a new framework which con-
sists of combining discretization and filter
methods. This framework is successfully ap-
plied to intrusion detection and microarray data
classification.
(b) Development of a novel method for dealing
with high-dimensional data: an ensemble of fil-
ters and classifiers. The idea of this ensemble is
to apply several filters based on different met-
rics and then joining the results obtained after
training a classifier with the selected subset of
features. In this manner, the user is released
from the task of choosing an adequate filter for
each dataset.
(c) Proposal for a new framework for cost-based
feature selection. In this manner, the scope of
feature selection is broaden by taking into con-
sideration not only the relevance of the features
but also their associated costs. The proposed
framework consists of adding a new term to the
evaluation function of a filter method so that the
cost is taken into account.
(d) Distributing feature selection. There are two
common types of data distribution: (a) horizon-
tal distribution wherein data are distributed in
subsets of instances; and (b) vertical distribu-
tion wherein data are distributed in subsets of
attributes. Both approaches are tested, employ-
ing for this sake filter and wrapper methods.
4 STAGE OF THE RESEARCH
This research has been started in 2009 and at present
if faces its final stage. As can be seen in Figure 1, the
great majority of objectives have been completed. For
the analysis of existing feature selection methods, the
first three tasks have been addressed. An investigation
about the particularities of classical methods has been
carried out, allowing to select the most appropriate
algorithms to face problems of real systems. More-
over, a study in detail about feature selection in mi-
croarray DNA data has been accomplished, since this
is a challenging domain which is a trending topic for
machine learning researchers. With the appearance
of high-dimensionality, it is also necessary to study
the scalability of existing feature selection methods.
The scalability of filter-based methods has been tack-
led with successful results. However, the analysis of
embedded and wrapper methods is still in progress
and some conclusions on this issue are expected to be
obtained soon.
The second objective of the thesis is also almost
completed. As mentioned in Section 1, the current
tendency in feature selection is not toward developing
new algorithmic measures, but toward favoring the
combination or modification of existing algorithms.
For this reason, this goal is focused in exploring dif-
ferent strategies to deal with the new problematics
which have emerged derived from the big data explo-
sion. Particularly, a novel method which combines
discretization and filter algorithms has been proposed
and its effectiveness was demonstratedin different ap-
plications, such as intrusion detection or microarray
data. Then, based on the assumption that a set of ex-
perts is better than a single expert, an ensemble of fil-
ters and classifiers was proposed and tested again on
microarray data as well as in some classical datasets.
Another task that has been concluded was to design
methods for cost-based feature selection, which has
been motivated by the fact that, in some cases, fea-
tures has its own risk or cost, and this factor must be
taken into account as well as the accuracy. Finally,
a recent topic of interest has arisen which consists
of distributing the feature selection process. For this
sake, several approaches have been proposed, split-
ting the data both vertically and horizontally. How-
ever, in some cases the partitioning of the datasets can
introduce some redundancy among features. For solv-
ing this problem, new partitioning schemes are being
investigated, for example by dividing the features ac-
cording to some goodness measure. Moreover, a final
research line in progress is devoted to perform paral-
lel feature selection using a cluster computing frame-
work called Spark (Spark, nd). This distributed pro-
gramming model has been proposed to handle large-
scale data problems. However, most existing feature
selection techniques are designed to run in a central-
ized computing environment and their implementa-
tions have to be adapted to this new technology. By
using Spark, the final user will be released of the de-
NovelFeatureSelectionMethodsforHighDimensionalData
5

cision of how to distribute the data.
5 RESULTS
As stated above, this thesis is facing its final stage
and therefore the objectives outlined in Section 3 have
been addressed. In this section, some experimental re-
sults as well as the key publications will be presented.
5.1 Review of Feature Selection
Methods
The first step when dealing with feature selection
should be to review the existing algorithms and to
check their performance under different situations. In
(Bol´on-Canedo et al., 2013d) a review of 11 classical
feature selection methods were applied over 11 syn-
thetic and 2 real datasets was presented. The main
objective of this work is to provide the user some rec-
ommendations about which feature selection method
is the most appropriate under a given type of data.
The suite of synthetic datasets chosen covers phe-
nomena such as the presence of irrelevant and redun-
dant features, noise in the data or interaction between
attributes. A scenario with a small ratio between num-
ber of samples and features where most of the features
are irrelevant was also tested. It reflects the prob-
lematic of datasets such as microarray data, a well-
known and hard challenge in the machine learning
field where feature selection becomes indispensable.
Within the feature selection field, three major ap-
proaches were evaluated: filters (correlation-based
feature selection – CFS, consistency-based, INTER-
ACT, Information Gain, ReliefF, minimum Redun-
dancy Maximum Relevance – mRMR and M
d
), wrap-
pers (with a support vector machine – SVM and a
C4.5 tree) and embedded methods (SVM recursive
feature elimination – SVM-RFE and feature selec-
tion perceptron – FS-P). To test the effectiveness of
the studied methods, an evaluation measure was in-
troduced trying to reward the selection of the relevant
features and to penalize the inclusion of the irrelevant
ones. Besides, four classifiers were selected (C4.5,
SVM, IB1 and naive Bayes) to measure the effective-
ness of the selected features and to check if the true
model was also unique.
Table 1 shows the behavior of the different feature
selection methods over the different problems stud-
ied, where the larger the number of dots, the better
the behavior. To decide which methods were the most
suitable under a given situation, it was computed a
trade-off between the proposed index of success and
the classification accuracy. In light of these results,
ReliefF turned out to be the best option independently
of the particulars of the data, with the added benefit
that it is a filter, which is the model with the low-
est computational cost. However, SVM-RFE with
a non-linear kernel showed outstanding results, al-
though its computational time is in some cases pro-
hibitive (in fact, it could not be applied over some
datasets). Wrappers have proven to be an interesting
choice in some domains, nevertheless they must be
applied together with their own classifiers and it has
to be reminded that this is the model with the highest
computational cost. In addition to this, Table 1 pro-
vides some guidelines for specific problems.
Table 1: Summary.
Method Correlation & Non Noise Noise No. feat >>
redundancy Linearity Inputs Target No. samples
CFS • • • ••• ••••
Consistency • • • ••• ••
INTERACT • • • ••• •••
InfoGain • • • ••• •••
ReliefF •••• ••••• ••••• ••••• ••
mRMR •••• ••• ••••• •• •
M
d
•••• •• ••• ••• •••
SVM-RFE •••• • • •••• •••••
SVM-RFEnl •••• ••••• ••• •••• –
FS-P ••••• •• •••• •••• •
Wrapper SVM • • ••• •••• ••
Wrapper C4.5 •• • •• ••• ••• •••
The feature selection methods were also tested
over two real datasets, demonstrating the conclusions
extracted from this theoretical study over real scenar-
ios, and proving the effectiveness of feature selection.
A preliminary study on this topic was published in
(Bol´on-Canedo et al., 2011c).
5.2 Application of Feature Selection to
Real Problems
After reviewing the behavior of the most famous fea-
ture selection methods over synthetic datasets, it is
necessary to prove their benefits on real problems.
This section will present real applications of this dis-
cipline, reporting success in different domains such
as classification of the tear film lipid layer and the K-
complex classification.
In (Bol´on-Canedo et al., 2012; Remeseiro et al.,
2013) a fast and automatic tool is presented to classify
the tear film lipid layer. The time required by previ-
ous approaches prevented their clinical use because it
was too long to allow the software tool to work in real
time. To solve this problem, feature selection plays a
crucial role since it reduces the number of input fea-
tures and, consequently, the processing time. Three
of the most popular feature selection methods were
chosen for this research: CFS, Consistency-based and
INTERACT. Those methods were tested over the fea-
ICAART2014-DoctoralConsortium
6

tures extracted from the images using co-occurence
features, a popular texture analysis method, in the
Lab colour space. Results showed that the CFS filter
surpass previous results in terms of processing time
whilst maintaining classification accuracy. In clin-
ical terms, the manual process done by experts can
be now automatized with the benefits of being faster,
with maximum accuracy over 96% and with a pro-
cessing time under 1 second. The clinical significance
of these results should be highlighted, as the agree-
ment between subjective observers is between 91%-
100%. Thus, it is completely recommended the use
of this application for clinical purposes as a support-
ing tool to diagnose evaporative dry eye.
The second real scenario was the K-complex clas-
sification (Hern´andez-Pereira et al., 2014), a key as-
pect in sleep studies. The same three filter methods
were applied combined with five different machine
learning algorithms, trying to achieve a low false pos-
itive rate whilst maintaining the accuracy. When fea-
ture selection was applied, the results improved sig-
nificantly for all the classifiers. It is remarkable the
91.40% of classification accuracy obtained by CFS,
reducing in 64% the number of features.
Notice that both problems are within the medi-
cal field, and in both cases the experts can take ad-
vantage of the feature selection. Not only are they
benefited from improvements in classification accu-
racy, but also from the model simplification, leading
in some cases to a better understanding of it.
5.3 Feature Selection on DNA
Microarray Classification
Among the different problems which have been
brought with the explosion of high-dimensional data,
one of the most important and studied is the analysis
of DNA microarray data. The key point to understand
all the attention devoted to this field is the challenge
that their problematic poses. Besides the obvious dis-
advantage of having so much features for such a small
number of samples, researchers have to deal also with
classes which are very unbalanced, training and test
datasets extracted under different conditions, dataset
shift or the presence of outliers. This is the reason be-
cause new methods emerge every year, not only trying
to improve previous results in terms of classification
accuracy, but also aiming to help biologists to iden-
tify the underlying mechanism that relates gene ex-
pression to diseases.
The research presented in (Bol´on-Canedo et al.,
2013) reviews the up-to-date contributions of feature
selection research applied to DNA microarray anal-
ysis, as well as the datasets used. Since the infancy
of microarray data classification, feature selection be-
came an imperative step, in order to reduce the num-
ber of features (genes).
Since the end of the nineties, when microarray
datasets began to be dealt with, a large number of
feature selection methods were applied. In the liter-
ature one can find both classical methods and meth-
ods developed especially for this kind of data. Due to
the high computational resources that these datasets
demand, wrapper and embedded methods have been
mostly avoided, in favor of less expensive approaches
such as filters.
The recent literature has been analyzed in order to
give the reader a brushstroke about the tendency in
developing feature selection methods for microarray
data. Furthermore, a summary of the datasets used in
the last years is provided. In order to have a complete
picture on the topic, we have also mentioned the most
common validation techniques. Since there is no con-
sensus in the literature about this issue, we have pro-
vided some guidelines.
Finally, a framework for feature selection evalu-
ation in microarray datasets has been proposed and
a practical evaluation where the results obtained are
analyzed. This experimental study tries to show in
practice the problematics that have been explained in
theory. For this sake, a suite of 9 widely-used bi-
nary datasets was chosen to apply over them 7 classi-
cal feature selection methods. For obtaining the final
classification accuracy, 3 well-known classifiers were
used. This large set of experiments aims also at facil-
itating future comparative studies when a researcher
proposes a new method.
Regarding the opportunities for future feature se-
lection research, the tendency is toward focusing on
new combinations such as hybrid or ensemble meth-
ods. This type of methods are able to enhance the
stability of the final subset of selected features, which
is also a trending topic in this domain. Another in-
teresting line of future research might be to distribute
the microarray data vertically (i.e. by features) in or-
der to reduce the heavy computational burden when
applying wrapper methods.
5.4 Scalability of Feature Selection
Methods
When dealing with the performanceof machine learn-
ing algorithms, most papers are focused on the accu-
racy obtained by the algorithm. However, with the
advent of high dimensionality problems, researchers
must study not only accuracy but also scalability.
Aiming at dealing with a problem as large as possible,
feature selection can be helpful as it reduces the input
NovelFeatureSelectionMethodsforHighDimensionalData
7

dimensionality and therefore the run-time required by
an algorithm.
In (Bol´on-Canedo et al., 2011a; Peteiro-Barral
et al., 2013) the effectiveness of feature selection on
the scalability of training algorithms for artificial neu-
ral networks (ANNs) was evaluated, both for classifi-
cation and regression tasks. Since there are no stan-
dard measures of scalability, those defined in the PAS-
CAL Large Scale Learning Challenge (Sonnenburg
et al., 2009) were used to assess the scalability of the
algorithms in terms of error, computational effort, al-
located memory and training time. Results showed
that feature selection as a preprocessing step is bene-
ficial for the scalability of ANNs, even allowing cer-
tain algorithms to be able to train on some datasets in
cases where it was impossible due to the spatial com-
plexity. Moreover, some conclusions about the ade-
quacy of the different feature selection methods over
this problem were extracted.
The next step was to evaluate the scalability of
the feature selection methods without the influence
of machine learning methods. An algorithm is said
to be scalable if it is suitable, efficient and practical
when applied to large datasets. However, the cur-
rent state is that the issue of scalability is far from
being solved although is present in a diverse set of
problems. Research on this topic has been collected
in (Peteiro-Barral et al., 2012; Bol´on-Canedo et al.,
2013; Rego-Fern´andez et al., 2014). An analysis of
the scalability of feature selection methods, which has
not received much consideration in the literature, has
been presented. Eight well-known filter-based feature
selection algorithms were evaluated, covering both
ranking and subset methods. A suite of ten artificial
datasets was chosen, so as to be able to assess the de-
gree of closeness to the optimal solution in a confident
way. For determining the scalability of the methods,
several new measures are proposed, based not only
in accuracy but also in execution time and stability,
and their adequacy was demonstrated. In light of the
experimental results, the fast correlation-based filter
(FCBF) seems to be the most scalable subset filter.
As for the ranker methods, ReliefF is a good choice
when having a small number of features (up to 128)
at the expense of a long training time. For this reason,
when dealing with extremely-high datasets, Informa-
tion Gain demonstrated better scalability properties.
5.5 Combination of Discretization and
Filter
The KDD (Knowledge Discovery and Data Mining
Tools Conference) Cup 99 dataset is a well-known
benchmark dataset with 5 million samples and 41 fea-
tures, which can be regarded as a multiclass or bi-
nary problem. Since some of its features are very
unbalanced, discretization is necessary prior to fea-
ture selection. A method based on the combination
of discretization, filtering and classification methods
that maintains the performance results using a re-
duced set of features is published in (Bol´on-Canedo
et al., 2011b). The results obtained in the binary
approach (Bol´on-Canedo et al., 2009; Bol´on-Canedo
et al., 2010b) outperformed the KDD Cup 99 compe-
tition winner result in performance, while using only
17% of the total number of features. Also, the KDD
Cup 99 dataset has been studied as a multiple class
problem, distinguishing among normal connections
and four types of attacks. Multiple class problems can
be dealt with by means of two different approaches:
using a multiple class algorithm and using multiple
binary classifiers. Both approaches were tested, but
with the first one, none of the results achieved im-
proved those obtained by the KDD winner.
For the multiple binary classifiers approach, two
class binarization techniques were utilized, namely
One vs Rest and One vs One. One of the results ob-
tained by One vs Rest and eight of the results obtained
by One vs One got a better score than the KDD win-
ner, so if the results of this research were in the orig-
inal contest, the KDD winner will be the tenth entry
of the competition. It is specially important the result
obtained with the One vs One approach, combining
the Proportional k-Interval Discretization (PKID) dis-
cretizer, the Consistency-based filter, the C4.5 classi-
fier and the Accumulative Sum decoding technique.
This result achieved a score of 0.2132 with only a
third of the input features, that improves the KDD
winner score in 0.0199. It is necessary to bear in mind
that the difference between the winner and the second
was only 0.0025.
This combination of discretizator and filter was
also applied successfully to several multiclass prob-
lems (S´anchez-Maro˜noet al., 2010) and to gene selec-
tion of microarray data (Bol´on-Canedo et al., 2010a;
Porto-D´ıaz et al., 2011). Remind that DNA microar-
ray data is a hard challenge for machine learning re-
searchers due to the high number of features (around
10 000) but small sample size (typically one hundred
or less).
5.6 An Ensemble of Filters and
Classifiers
There is a vast body of feature selection methods
in the literature, based on different metrics, and to
choose the adequate method for each scenario is not
an easy-to-solvequestion. The proliferation of feature
ICAART2014-DoctoralConsortium
8
selection algorithms has not brought about a general
methodology that allows for intelligent selection from
existing algorithms. For a specific dataset, employing
one or another feature selection method varies the se-
lected subset of features and, consequently, the per-
formance result obtained by a machine learning algo-
rithm. In order to reduce the variability associated to
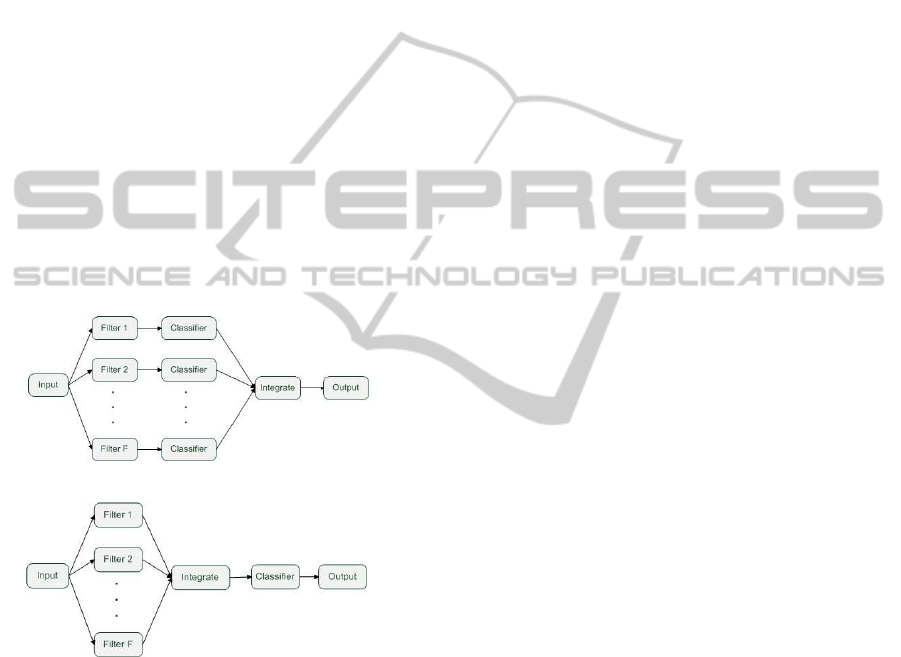
feature selection, an ensemble of filters has been pro-
posed and published in (Bol´on-Canedo et al., 2012)
in order to obtain good performanceindependently on
the dataset. The idea was to combine several filters,
employing different metrics and performing a feature
reduction. Each filter selects a subset of features and
this subset is used for training the classifier. There
will be as many outputs as filters employed and the re-
sult of the filters and classifier will be combined using
simple voting (see Figure 2(a)). The experimental re-
sults on DNA microarray data showed that, although
in some specific cases there is a filter that performs
better than the ensemble, there is not a better filter in
general, and the ensemble seems to be the most reli-
able alternative when a feature selection process has
to be carried out.
(a) Ensemble 1
(b) Ensemble 2
Figure 2: Implementations of the ensemble.
Then, the previous study was extended with an-
other approach (Bol´on-Canedo et al., 2011d; Bol´on-
Canedo et al., 2013a), changing the role of the clas-
sifier. Ensemble1 (see Figure 2(a)) classifies as many
times as there are filters, whereas Ensemble2 (see Fig-
ure 2(b)) classifies only once with the result of join-
ing the different subsets selected by the filters. For
Ensemble1, two methods for combining the outputs
of the classifiers were studied, as well as the possibil-
ity of using an adequate specific classifier for each
filter. A total of five different implementations of
the two approaches of ensemble were proposed: E1-
sv, which is Ensemble1 using simple voting as com-
bination method; E1-cp, which is Ensemble1 using
cumulative probabilities as combination method; E1-
ni, which is Ensemble1 with specific classifiers naive
Bayes and IB1; E1-ns, which is Ensemble1 with spe-
cific classifiers naive Bayes and SVM and E2, which
is Ensemble2.
Results over synthetic data showed the adequacy
of the proposed methods on this controlled scenario
since they selected the correct features. The next
step was to apply these approaches to 5 UCI classical
datasets. Experimental results demonstrated that one
of the ensembles (E1-cp) combined with C4.5 classi-
fier was the best option when dealing with this type
of dataset. Finally, the ensemble configurations were
tested over 7 DNA microarray data. As expected, us-
ing an ensemble was again the best option. Specifi-
cally, the best performance was achieved again with
E1-cp but this time combined with SVM classifier. It
should be noted that some of these datasets presented
a high imbalance of the data. To overcome this prob-
lem, an oversampling method was applied after the
feature selection process. The result was that once
again one of the ensembles achieved the best perfor-
mance, and that this was even better than the one ob-
tained with no preprocessing, showing the adequacy
of the ensemble combined with over-sampling meth-
ods. Thus, the appropriateness of using an ensemble
instead of a single filter remained demonstrated, con-
sidering that for all scenarios tested, the ensemble was
always the more successful solution.
Regarding the different implementations of the
ensemble tested, several conclusions can be drawn.
There is a slight difference between the two combiner
methods employed with Ensemble1 (simple voting
and cumulative probability), although the second one
obtained the best performance. Among the different
classifiers chosen for this study, it appeared that the
type of data to be classified determines significantly
the error achieved, so it is responsibility of the user to
know which classifier is more suitable for a given type
of data. The authors recommend using E1-cp with
C4.5 when classifying classical datasets (with more
samples than features) and E1-cp with SVM when
dealing with microarray dataset (with more features
than samples). In complete ignorance of the partic-
ulars of the data, we suggest using E1-ns, which re-
leases the user from the task of choosing a specific
classifier.
5.7 Cost-based Feature Selection
There is a broad suite of filter methods, based on dif-
ferent metrics, but the most common approaches are
to find either a subset of features that maximizes a
NovelFeatureSelectionMethodsforHighDimensionalData
9

given metric or either an ordered ranking of the fea-
tures based on this metric. However, there are some
situations where a user is not only interested in max-
imizing the merit of a subset of features, but also in
reducing costs that may be associated to features. For
example, for medical diagnosis, symptoms observed
with the naked eye are costless, but each diagnostic
value extracted by a clinical test is associated with its
own cost and risk. In other fields, such as image anal-
ysis, the computational expense of features refers to
the time and space complexities of the feature acqui-
sition process. This is a critical issue, specifically in
real-time applications, where the computational time
required to deal with one or another feature is cru-
cial (see Section 5.2), and also in the medical domain,
where it is important to save economic costs and to
also improve the comfort of a patient by preventing
risky or unpleasant clinical tests (variables that can be
also treated as costs).
In (Bol´on-Canedo et al., 2014) a new framework
for cost-based feature selection is proposed. The ob-
jective is to solve problems where not only it is in-
teresting to minimize the classification error, but also
reducing costs that may be associated to input fea-
tures. This framework consists of adding a new term
to the evaluation function of any filter feature selec-
tion method so that it is possible to reach a trade-off
between a filter metric (e.g. correlation or mutual in-
formation) and the cost associated to the selected fea-
tures. A new parameter, called λ, is introduced in or-
der to adjust the influence of the cost into the eval-
uation function, allowing the user fine control of the
process according to his needs.
In order to test the adequacy of the proposed
framework, two well-known and representative filters
are chosen: CFS (belonging to the subset feature se-
lection methods) and mRMR (belonging to the ranker
feature selection methods). Experimentation is exe-
cuted over a broad suite of different datasets. Results
after performing classification with a SVM display
that the approach is sound and allow the user to re-
duce the cost without compromising the classification
error significantly, which can be very useful in fields
such as medical diagnosis or real-time applications.
Then, in (Bol´on-Canedo et al., 2014), a modifi-
cation of the ReliefF filter for cost-based feature se-
lection, called mC-ReliefF, is proposed. Twelve dif-
ferent datasets, covering very diverse situations, were
selected to test the approach. Results after performing
classification with a SVM and Kruskal-Wallis statisti-
cal tests, again demonstrated the adequacy of the cost-
based feature selection. Finally, the method was ap-
plied to the real problem presented in Section 5.2: the
tear film lipid layer classification. In this scenario the
time required to extract the features prevented clinical
use because it was too long to allow the software tool
to work in real time. mC-ReliefF permits to automat-
ically decrease the required time (from 38 seconds to
less than 1 second, that is in 92%) while maintaining
the classification performance. Notice that this reduc-
tion in time is very important since interviews with
optometrists revealed that a scale of computation time
over 10 seconds per image makes the system not us-
able.
5.8 Distributed Feature Selection
Traditionally, feature selection methods are applied in
a centralized manner, i.e. a single learning model to
solve a given problem. However, when dealing with
large amounts of data, distributed feature selection
seems to be a promising line of research since allocat-
ing the learning process among several workstations
is a natural way of scaling up learning algorithms.
Moreover, it allows to deal with datasets that are nat-
urally distributed, a frequent situation in many real
applications (e.g. weather databases, financial data
or medical records). There are two common types of
data distribution: (a) horizontal distribution wherein
data are distributed in subsets of instances; and (b)
vertical distribution wherein data are distributed in
subsets of attributes.
The great majority of approaches distribute the
data horizontally, since it constitutes the most suit-
able and natural approach for most applications. In
(Bol´on-Canedo et al., 2013), a methodology is pro-
posed which consists of applying filters over several
partitions of the data, combined in the final step into a
single subset of features. The idea of distributing the
data horizontally builds on the assumption that com-
bining the output of multiple experts is better than the
output of any single expert. There are three main
stages: (i) partition of the datasets; (ii) application
of the filter to the subsets; and (iii) combination of
the results. An experimental study was carried out
on six datasets considered representative of problems
from medium to large size. In terms of classifica-
tion accuracy, our distributed filtering approach ob-
tains similar results to the centralized methods, even
with slight improvements for some datasets. Further-
more, the most important advantage of the proposed
method is the dramatically reduction in computational
time (from the order of hours to the order of minutes).
While not common, there are some other develop-
ments that distribute the data by features. In (Bol´on-
Canedo et al., 2013b) the data are distributed verti-
cally in order to have the feature selection process dis-
tributed. This approach is especially suitable for mi-
ICAART2014-DoctoralConsortium
10
croarray data since in this manner we will deal with
subsets with a more balanced features/samples ratio
and avoid overfitting problems.
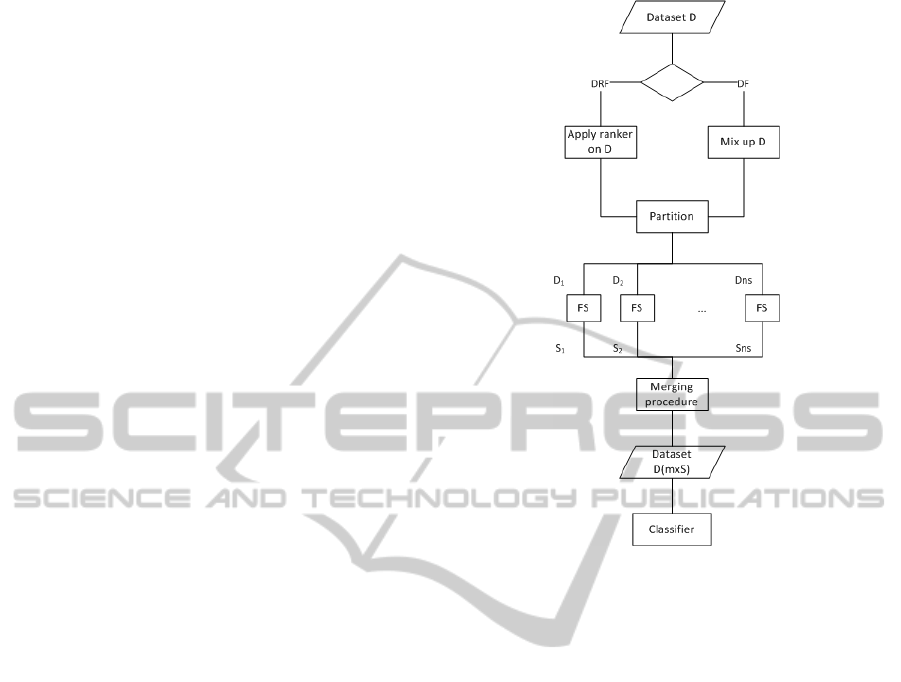
The partition of the dataset consists of dividing
the original dataset into several disjoint subsets of ap-
proximately the same size that cover the full dataset.
Two different methods were used for partitioning the
data: (a) performing a randomly partition and (b)
ranking the original features before generating the
subsets. The second option was introduced trying to
improve the performance results obtained by the first
one. By having an ordered ranking, features with sim-
ilar relevance to the class will be in the same sub-
set, which will facilitate the task of the subset filter
which will be applied later. These two techniques for
partitioning the data will generate two different ap-
proaches for the distributed method: Distributed Fil-
ter (DF) with the randomly partition and Distributed
Ranking Filter (DRF) associated to the ranking parti-
tion.
After this step, the data is split by assigning groups
of k features to each subset, where the number of fea-
tures k in each subset is half the number of samples, to
avoid overfitting. When opting for the randomly par-
tition (DF), the groups of k features are constructed
randomly, having into account that the subsets have
to be disjoint. In the case of the ranking partition
(DRF), the groups of k features are generated sequen-
tially over the ranking, so features with a similar rank-
ing position will be in the same group. Notice that
the random partition is equivalent to obtain a random
ranking of the features and then follow the same steps
as with the ordered ranking. Figure 3 shows a flow
chart which reflects the two algorithms proposed, DF
and DRF. After having several small disjoint datasets
D
i
, the filter method will be applied to each of them,
returning a selection S
i
for each subset of data. Fi-
nally, to combine the results, a merging procedure us-
ing a classifier will be executed.
The experiments on eight microarray datasets
showed that this proposal was able to reduce the run-
ning time significantly with respect to the standard
(centralized) filtering algorithms. In terms of execu-
tion time, the behavior is excellent, being this fact
the most important advantage of our method. Fur-
thermore, with regard of classification accuracy, our
distributed approach was able to match and in some
cases even improve the standard algorithms applied
to the non-partitioned datasets. This situation is re-
flected in Figure 4, where the best result among all
the classifiers is displayed for any dataset and the
consistency-based filter. It is easy to see at a glance
that the accuracies fall into similar values (being most
of the time a distributed approach the best option)
Figure 3: Flow chart of distributed filter approach.
whilst the differences in time are outstanding.
Finally, in (Bol´on-Canedo et al., 2013c), the ad-
equacy of a distributed approach for wrapper fea-
ture selection was tested over four datasets consid-
ered representative of problems from medium to large
size. The goal was to design a distributed wrapper
which would led to a reduction in the running time
as well as in the storage requirements while the accu-
racy would not drop to inadmissible values. Again,
the experiments showed that our method was able to
shorten the execution time impressively compared to
the standard wrapper algorithms. Furthermore, our
distributed wrapper achieved a similar performance
to the original wrapper. In terms of test accuracy, the
proposed distributed wrapper is able to match and in
some cases even to improve the standard results ap-
plied to the non-partitioned datasets.
6 CONCLUSIONS
Continual advances in computer-based technologies
have enabled researchers and engineers to collect data
at an increasingly fast pace. To address this challenge,
feature selection becomes an imperative preprocess-
ing step which needs to be adapted and improved to
handle high-dimensional data.
This work is devoted to study feature selection and
NovelFeatureSelectionMethodsforHighDimensionalData
11
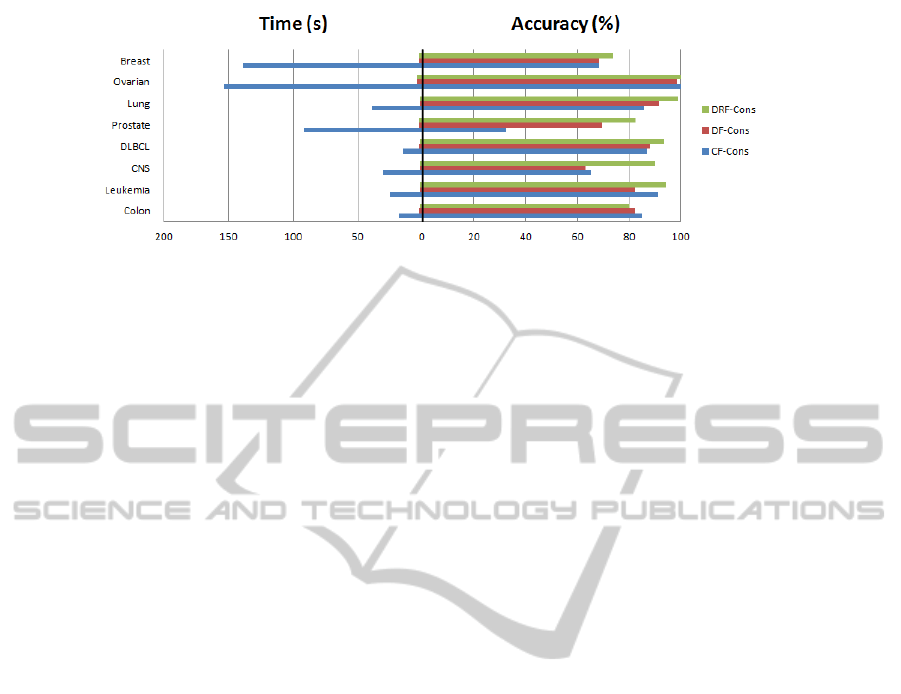
Figure 4: Comparison of accuracy and time for consistency-based filter.
its adequacy to large-scale data. The tendency nowa-
days is two-fold: on the one hand, to improve and
extend the existing methods to address the new chal-
lenges associated to high-dimensionality. And, on the
other hand, to develop novel techniques to directly
solving the arising challenges.
First, a critical analysis of existing feature selec-
tion was performed, to check their adequacy toward
different challenges and be able to provide some rec-
ommendations. Bearing this analysis in mind, the
most adequate techniques were applied to several
real-life problems, obtaining a notable improvement
in performance. Apart from efficiency, another crit-
ical issue in large-scale applications which is scala-
bility. The effectiveness of feature selection methods
may be significantly downgraded, if not totally inap-
plicable, when the data size increases steadily. For
this reason, a stability analysis in detail of the most
famous techniques was done.
Then, new techniques for large-scale feature se-
lection were proposed. In the first place, as most
of the existing feature selection techniques need data
to be discrete, a new approach was proposed that
consists in a combination of a discretizer, a filter
method and a very simple classical classifier, obtain-
ing promising results. Another proposal was to em-
ploy a ensemble of filters instead of a single one, re-
leasing the user from the decision of which technique
is the most appropriate for a given problem. An in-
teresting topic is also to consider the cost related with
the different features, therefore a framework for cost-
based feature selection was proposed, demonstrating
its adequacy in a real-life scenario. Finally, it is well-
known that a manner of handling large-scale data is to
transform the large-scale problem into several small-
scale problems, by distributing the data. With this
aim, several approaches for distributed and parallel
feature selection have been proposed.
As can be seen, this thesis covers a broad
suite of problems arisen from the advent of high-
dimensionality. The proposed approaches have
demonstrated to be sound, and it is expected that their
contribution will be important in the next years, since
feature selection for large-scale data is likely to con-
tinue to be a trending topic in the near future.
ACKNOWLEDGEMENTS
This research has been partially funded by the Secre-
tar´ıa de Estado de Investigaci´on of the Spanish Gov-
ernment and FEDER funds of the European Union
through the research projects TIN 2012-37954 and
PI10/00578; and by the Conseller´ıa de Industria of
the Xunta de Galicia through the research projects
CN2011/007 and CN2012/211. Veronica Bolon-
Canedo acknowledges the support of Xunta de Gali-
cia under Plan I2C Grant Program.
REFERENCES
Bol´on-Canedo, V., Peteiro-Barral, D., Alonso-Betanzos,
A., Guijarro-Berdi˜nas, B., and S´anchez-Maro˜no, N.
(2011a). Scalability analysis of ann training algo-
rithms with feature selection. In Advances in Artificial
Intelligence, pages 84–93. Springer.
Bol´on-Canedo, V., Peteiro-Barral, D., Remeseiro, B.,
Alonso-Betanzos, A., Guijarro-Berdinas, B., Mos-
quera, A., Penedo, M. G., and S´anchez-Maro˜no, N.
(2012). Interferential tear film lipid layer classifica-
tion: an automatic dry eye test. In Tools with Artificial
Intelligence (ICTAI), 2012 IEEE 24th International
Conference on, volume 1, pages 359–366. IEEE.
Bol´on-Canedo, V., Porto-D´ıaz, I., S´anchez-Maro˜no, N.,
and Alonso-Betanzos, A. (2014). A framework for
cost-based feature selection. Pattern Recognition (In
press).
Bol´on-Canedo, V., Rego-Fern´andez, D., Peteiro-Barral,
D., Alonso-Betanzos, A., Guijarro-Berdinas, B., and
S´anchez-Maro˜no, N. (2013). On the scalability of fil-
ter techniques for feature selection on big data. IEEE
Computational Intelligence Magazine Special Issue
on Computational Intelligence in Big Data (Under Re-
view).
ICAART2014-DoctoralConsortium
12

Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2009). A combination of discretiza-
tion and filter methods for improving classification
performance in kdd cup 99 dataset. In Neural Net-
works, 2009. IJCNN 2009. International Joint Con-
ference on, pages 359–366. IEEE.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2010a). On the effectiveness of dis-
cretization on gene selection of microarray data. In
International Joint Conference on Neural Networks.
IJCNN 2010, pages 3167–3174. IEEE.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2011b). Feature selection and classi-
fication in multiple class datasets: An application to
kdd cup 99 dataset. Expert Systems with Applications,
38(5):5947–5957.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2011c). On the behavior of feature se-
lection methods dealing with noise and relevance over
synthetic scenarios. In Neural Networks (IJCNN), The
2011 International Joint Conference on, pages 1530–
1537. IEEE.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2011d). Toward an ensemble of filters
for classification. In Intelligent Systems Design and
Applications (ISDA), 2011 11th International Confer-
ence on, pages 331–336. IEEE.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2012). An ensemble of filters and classi-
fiers for microarray data classification. Pattern Recog-
nition, 45(1):531–539.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2013a). Data classification using an en-
semble of filters. Neurocomputing (In Press).
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2013b). A distributed filter approach
for microarray data classification. Applied Soft Com-
puting (Under Review).
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2013c). A distributed wrapper approach
for feature selection. In European Symposium on Arti-
ficial Neural Networks, ESANN 2013, pages 173–178.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2013d). A review of feature selection
methods on synthetic data. Knowledge and informa-
tion systems, 34(3):483–519.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Alonso-
Betanzos, A. (2014). mc-relieff: An extension of
relieff for cost-based feature selection. In 6th Inter-
national Conference on Agents and Artificial Intelli-
gence (ICAART) (Accepted).
Bol´on-Canedo, V., S´anchez-Maro˜no, N., Alonso-Betanzos,
A., Ben´ıtez, J., and Herrera, F. (2013). An insight into
microarray datasets and feature selection methods: a
framework for ongoing studies. Information Sciences
(Under review).
Bol´on-Canedo, V., S´anchez-Maro˜no, N., Alonso-Betanzos,
A., and Hernandez-Pereira, E. (2010b). Feature selec-
tion and conversion methods in KDD Cup 99 dataset:
A comparison of performance. In Proceedings of the
10th IASTED International Conference, pages 58–66.
Bol´on-Canedo, V., S´anchez-Maro˜no, N., and Cervi˜no-
Rabu˜nal, J. (2013). Scaling up feature selection: A
distributed filter approach. In Advances in Artificial
Intelligence, pages 121–130. Springer.
Chidlovskii, B. and Lecerf, L. (2008). Scalable feature se-
lection for multi-class problems. In Machine Learning
and Knowledge Discovery in Databases, pages 227–
240. Springer.
Dash, M. and Liu, H. (1997). Feature selection for classifi-
cation. Intelligent data analysis, 1(3):131–156.
Frank, A. and Asuncion, A. (2010). UCI Machine Learn-
ing Repository. http://archive.ics.uci.edu/ml. [Online;
accessed December-2013].
Guyon, I., Gunn, S., Nikravesh, M., and Zadeh, L. (2006).
Feature extraction: foundations and applications, vol-
ume 207. Springer.
Hern´andez-Pereira, E., Bol´on-Canedo, V., S´anchez-
Maro˜no, N.,
´
Alvarez-Est´evez, D., Moret-Bonillo, V.,
and Alonso-Betanzos, A. (2014). A comparison of
performance of k-complex classification methods us-
ing feature selection. Information Sciences (Under re-
view).
Hughes, G. (1968). On the mean accuracy of statistical pat-
tern recognizers. Information Theory, IEEE Transac-
tions on, 14(1):55–63.
Kohavi, R. and John, G. H. (1997). Wrappers for feature
subset selection. Artificial intelligence, 97(1):273–
324.
Liu, H. and Yu, L. (2005). Toward integrating feature
selection algorithms for classification and clustering.
Knowledge and Data Engineering, IEEE Transactions
on, 17(4):491–502.
Loscalzo, S., Yu, L., and Ding, C. (2009). Consensus
group stable feature selection. In Proceedings of
the 15th ACM SIGKDD international conference on
Knowledge discovery and data mining, pages 567–
576. ACM.
Peng, Y., Wu, Z., and Jiang, J. (2010). A novel feature
selection approach for biomedical data classification.
Journal of Biomedical Informatics, 43(1):15–23.
Peteiro-Barral, D., Bolon-Canedo, V., Alonso-Betanzos,
A., Guijarro-Berdinas, B., and S´anchez-Maro˜no, N.
(2012). Scalability analysis of filter-based methods
for feature selection. Advances in Smart Systems Re-
search, 2(1):21–26.
Peteiro-Barral, D., Bol´on-Canedo, V., Alonso-Betanzos,
A., Guijarro-Berdi˜nas, B., and S´anchez-Maro˜no, N.
(2013). Toward the scalability of neural networks
through feature selection. Expert Systems with Ap-
plications, 40(8):2807–2816.
Porto-D´ıaz, I., Bol´on-Canedo, V., Alonso-Betanzos, A.,
and Fontenla-Romero, O. (2011). A study of perfor-
mance on microarray data sets for a classifier based
on information theoretic learning. Neural Networks,
24(8):888–896.
Provost, F. (2000). Distributed data mining: Scaling up and
beyond. Advances in distributed and parallel knowl-
edge discovery, pages 3–27.
Rego-Fern´andez, D., Bol´on-Canedo, V., and Alonso-
Betanzos, A. (2014). Scalability analysis of mrmr
for microarray data. In 6th International Conference
NovelFeatureSelectionMethodsforHighDimensionalData
13

on Agents and Artificial Intelligence (ICAART) (Ac-
cepted).
Remeseiro, B., Bol´on-Canedo, V., Peteiro-Barral, D.,
Alonso-Betanzos, A., Guijarro-Berdinas, B., Mos-
quera, A., Penedo, M. G., and S´anchez-Maro˜no, N.
(2013). A methodology for improving tear film lipid
layer classification. IEEE Journal of Biomedical and
Health Informatics (In Press).
Saeys, Y., Abeel, T., and Van de Peer, Y. (2008). Robust fea-
ture selection using ensemble feature selection tech-
niques. In Machine Learning and Knowledge Discov-
ery in Databases, pages 313–325. Springer.
S´anchez-Maro˜no, N., Alonso-Betanzos, A., Garc´ıa-
Gonz´alez, P., and Bol´on-Canedo, V. (2010). Mul-
ticlass classifiers vs multiple binary classifiers using
filters for feature selection. In International Joint
Conference on Neural Networks. IJCNN 2010, pages
2836–2843. IEEE.
Sonnenburg, S., Franc, V., Yom-Tov, E., and Sebag, M.
(2009). PASCAL Large Scale Learning Challenge.
Journal of Machine Learning Research.
Spark (n.d.). Apache Spark - Lightning-Fast Cluster Com-
puting. http://spark.incubator.apache.org. [Online; ac-
cessed December-2013].
Sun, Y. and Li, J. (2006). Iterative relief for feature weight-
ing. In Proceedings of the 23rd international confer-
ence on Machine learning, pages 913–920. ACM.
Sun, Y., Todorovic, S., and Goodison, S. (2008). A feature
selection algorithm capable of handling extremely
large data dimensionality. In SDM, pages 530–540.
Tuv, E., Borisov, A., Runger, G., and Torkkola, K. (2009).
Feature selection with ensembles, artificial variables,
and redundancy elimination. The Journal of Machine
Learning Research, 10:1341–1366.
Vainer, I., Kraus, S., Kaminka, G. A., and Slovin, H. (2011).
Obtaining scalable and accurate classification in large-
scale spatio-temporal domains. Knowledge and infor-
mation systems, 29(3):527–564.
Yu, L. and Liu, H. (2004). Efficient feature selection via
analysis of relevance and redundancy. The Journal of
Machine Learning Research, 5:1205–1224.
Zhang, Y., Ding, C., and Li, T. (2008). Gene selection algo-
rithm by combining relieff and mrmr. BMC genomics,
9(Supl 2):S27.
Zhao, Z. and Liu, H. (2011). Spectral Feature Selection for
Data Mining. Chapman & Hall/Crc Data Mining and
Knowledge Discovery. Taylor & Francis Group.
ICAART2014-DoctoralConsortium
14
