
Modelling and Analysis of Retinal Ganglion Cells
Through System Identification
Dermot Kerr
1
, Martin McGinnity
2
and Sonya Coleman
1
1
School of Computing and Intelligent Systems, University of Ulster, Magee campus, Derry, Northern Ireland
2
School of Science & Technology, Nottingham Trent University, Nottingham, U.K.
Keywords: System Identification, Retinal Ganglion Cells, Linear-Nonlinear Model.
Abstract: Modelling biological systems is difficult due to insufficient knowledge about the internal components and
organisation, and the complexity of the interactions within the system. At cellular level existing
computational models of visual neurons can be derived by quantitatively fitting particular sets of
physiological data using an input-output analysis where a known input is given to the system and its output
is recorded. These models need to capture the full spatio-temporal description of neuron behaviour under
natural viewing conditions. At a computational level we aspire to take advantage of state-of-the-art
techniques to accurately model non-standard types of retinal ganglion cells. Using system identification
techniques to express the biological input-output coupling mathematically, and computational modelling
techniques to model highly complex neuronal structures, we will "identify" ganglion cell behaviour with
visual scenes, and represent the mapping between perception and response automatically.
1 INTRODUCTION
Modelling biological systems is difficult due to
insufficient knowledge about the internal
components and organisation, and the complexity of
the interactions within the system. System
identification has emerged as a viable alternative to
classical hypothesis testing for the understanding of
biological systems and was first used to understand
the responses of auditory neurons (De Boer, 1968).
Using white noise stimuli as input, the output
responses were recorded and inferences made on
mapping the stimulus to the response. White noise
stimulation is often selected to model biological
vision systems (Sakai, 1988, Chichilnisky, 2001) as
it is mathematically simple to analyse. However, it is
unlikely that white noise stimuli would test the full
function of a neuron’s behaviour (Talebi, 2012).
Thus, any model developed with this stimulus could
only be considered a subset of the biological model
under certain conditions.
In the work by Marmarelis (Marmarelis, 1972), the
Wiener theory of nonlinear system identification
was applied to study the underlying operation of the
three stage neuronal structures in the catfish retina.
Following from this work, the Volterra-Wiener
method has been used extensively to model
nonlinear biological systems (Victor, 1977, 1979,
Marmarelis, 2004, Korenberg, 1996). However,
computational effort increases geometrically with
the kernel order and in interpretation of higher order
kernels (Herikstad, 2011). Marmarelis and Zhao
(Marmarelis, 1997) presented a way of overcoming
these limitations by developing a perceptron type
network with polynomial activation functions.
Block-structured (Giri, 2010) or modular models
in the form of cascaded or parallel configurations
have been used to overcome the limitations of
Volterra-Wiener models. Cascade models may take
various forms such as linear-nonlinear (Ostojic,
2011), nonlinear-linear, linear-nonlinear-linear, etc.
In particular, linear-nonlinear models have been
used to describe the processing in the retina (Pillow,
2005).
The generalised modular model proposed by
Korenberg (Korenberg, 1991) employed parallel
linear-nonlinear cascades generating spike outputs
with a threshold-trigger function. To model specific
neuron responses such as burstiness, refractoriness
and gain control, Pillow (Pillow, 2008) amended the
linear-nonlinear models with feedback terms.
Correlated neuron activity was modelled through the
158
Kerr D., McGinnity M. and Coleman S..
Modelling and Analysis of Retinal Ganglion Cells Through System Identification.
DOI: 10.5220/0005069701580164
In Proceedings of the International Conference on Neural Computation Theory and Applications (NCTA-2014), pages 158-164
ISBN: 978-989-758-054-3
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

use of coupling filters (Pillow, 2008) to couple
multiple linear-nonlinear models of individual cells.
Parametric system identification modelling
techniques also exist. The NARMAX (nonlinear
auto-regressive moving average with exogenous
inputs) model (Billings, 1984) has been used to
model the nonlinear behaviour observed in the fly
photoreceptors (Friederich, 2009, Song, 2009). The
NARMAX modelling technique is suitable for
application in a number of areas and has also been
used to model robot behaviour (Kerr, 2010), iceberg
calving and detecting and tracking time-varying
causality for EEG data (Billings, 2013). Neural
network approaches have also been used to model
biological aspects of the vision system. For example
Lau (Lau, 2002) used a two layer neural network
with the backpropagation training algorithm to
model the nonlinear responses of neurons in the
visual cortex to visual stimuli. Similarly, Prenger
(Prenger, 2004) used a multilayer feed-forward
neural network to model the nonlinear stimulus-
response relationship in the primary visual cortex
using natural images.
In this paper we formalise and standardise the
model development process by using system
identification techniques (NARMAX modelling) to
express the biological input-output coupling
mathematically. We have used a NARMAX
approach to obtain the models we need because:
The NARMAX model itself provides the
executable code straight away,
The model is analysable, and gives us valuable
information regarding
How the model achieves the task,
Whether the model is stable or not,
How the model will behave under certain
operating conditions, and
How sensitive the model is to certain inputs,
i.e. how “important” certain input are.
In Section 2 we present the visual stimuli used in the
neuronal recordings to obtain the physiological data
and in Section 3 we present an overview of the
NARMAX modelling approach. Experiments and
results are presented in Section 4 with discussion in
Section 5.
2 NEURONAL DATA
Recordings were obtained from isolated mice retinas
under full field stimulation using a Gaussian white
noise sequence as illustrated in Figure 1.
Figure 1: Full-field Gaussian white noise sequence.
The isolated retina was placed on a multi-electrode
array, which recorded spike trains from many
ganglion cells simultaneously. Stimuli were
projected onto the isolated retina via a miniature
cathode ray tube monitor. Spikes were sorted off-
line by a cluster analysis of their shapes, and spike
times were measured relative to the beginning of
stimulus presentation. In the experiments presented
in this paper we analyse the response from an ON
retinal ganglion cell (RGC).
3 NARMAX MODELLING
The NARMAX model is a difference equation that
expresses the present value of the output as a
nonlinear combination of previous values of the
output, previous and/or present values of the input,
and previous and/or present values of the noise
signal. NARMAX is a parameter estimation
methodology for identifying both the important
model terms and the parameters of an unknown non-
linear dynamic system, such as a sensory neuron.
For single-input single-output systems this model
takes the form:
)(
)](,),1(
),(,),(
),(,),2(),1([)(
ke
nekeke
nudkudku
nykykykyFky
(1)
where )(ky , )(ku , )(ke are the sampled output,
input and unobservable noise sequences
respectively,
nenuny ,,
are the regression orders of
)(ky , )(ku , )(ke and d is a time delay. []F is a
nonlinear function and is typically taken to be a
polynomial expansion of the arguments. Usually
only the input and output measurements are
ModellingandAnalysisofRetinalGanglionCellsThroughSystemIdentification
159

available and the investigator must process these
signals to estimate a model of the system.
The NARMAX methodology divides this
problem into the following steps:
Structure detection;
Parameter estimation;
Model validation;
Prediction;
Analysis.
These steps form an estimation toolkit that allows
the user to build a concise mathematical description
of the system (Billings and Chen, 1998). The
procedure begins by determining the structure or the
important model terms using a special orthogonal
least squares procedure. This algorithm determines
which dynamic and nonlinear terms should be in the
model by computing the contribution that each
potential model term makes to the system output.
This allows the model to be built up term by term in
a manner that exposes the significance of each new
term that is added.
Structure detection is a fundamental part of
the NARMAX procedure because searching for the
structure ensures that the model is as simple as
possible and a model with good generalisation
properties is obtained. This approach mimics
analytical modelling methods where the important
model terms are introduced first. Subsequently the
model is refined by adding in less significant effects.
The only difference is that in the NARMAX method
the model terms can be identified from the data set.
These procedures are now well established
and have been used in many modelling domains.
Once the structure of the model has been determined
the unknown parameters in the model can be
estimated. If correct parameter estimates are to be
obtained the noise sequence, e(k) which is almost
always unobservable, must be estimated and
accommodated within the model. Model validation
methods are then applied to determine if the model
is adequate.
Once the model has been determined to be
adequate it can be used to predict the system output
for different inputs. The model may also be used to
study the characteristics of the system under
investigation (Nehmzow, 2006). It is this latter
aspect that is of particular interest in the work
presented here. In this paper we have examined the
suitability of NARMAX modelling to express the
biological stimulus-response coupling
mathematically and to validate the resulting
stimulus-response couplings.
4 MODEL IDENTIFICATION
PROCEDURE AND ANALYSIS
The proposed approach represents a decisive
departure from current methods of generating retina
models. We propose to "identify" (in the sense of
system identification) the neuron’s behaviour with
natural visual scenes, and to represent the mapping
between perception and response automatically,
using the NARMAX system identification
technique. We will illustrate how the various neural
networks within the layered retina structure can be
modelled using efficient polynomials that
incorporate the neuron's nonlinear behaviour and
dynamics. The compact polynomial representation
will demonstrate that the intricate retina neural
networks may be modelled in a compact compressed
form.
4.1 Data Pre-processing
The overall goal of the pre-processing stage is to
manipulate the data so that they form a regression
dataset, i.e. input-output corresponding to the
stimulus-response. In this case the dataset will be
single-input single-output.
The Gaussian white noise is a stochastic highly
interleaved stimuli spanning a wide range of visual
inputs, is relatively robust to fluctuations in
responsivity, avoids adaptation to strong or
prolonged stimuli and is well suited to simultaneous
measurements from multiple neurons. Examples of
stimuli are presented in Figure 1 where each image
in the sequence is presented sequentially to the
isolated retina. As the stimulus has uniform intensity
there is no need to extract the stimulus in the region
of the receptive field.
Recordings of the ganglion cell neural response
(spikes) to the full-field stimulation were supplied
for two different ganglion cells in the case of this
dataset. Each file contains the recorded times of
spikes in seconds. For example,
[1.76304, 1.76912, 1.78504,…,546.63776].
Using these recorded spike times we compute a
continuous temporal spike rate using the standard
method of binning and convolution with a window
function. Using this method we then have a
continuous valued input-output dataset. For
example, in Figure 2 we have illustrated the input
data (stimulus intensity), recorded spikes and
computed spike rate using an alpha window function
for 500 milliseconds of a recording. Figure 2(a)
illustrates the stimulus intensity, Figure 2(b)
illustrates the recorded spikes, and Figure 2(c)
NCTA2014-InternationalConferenceonNeuralComputationTheoryandApplications
160
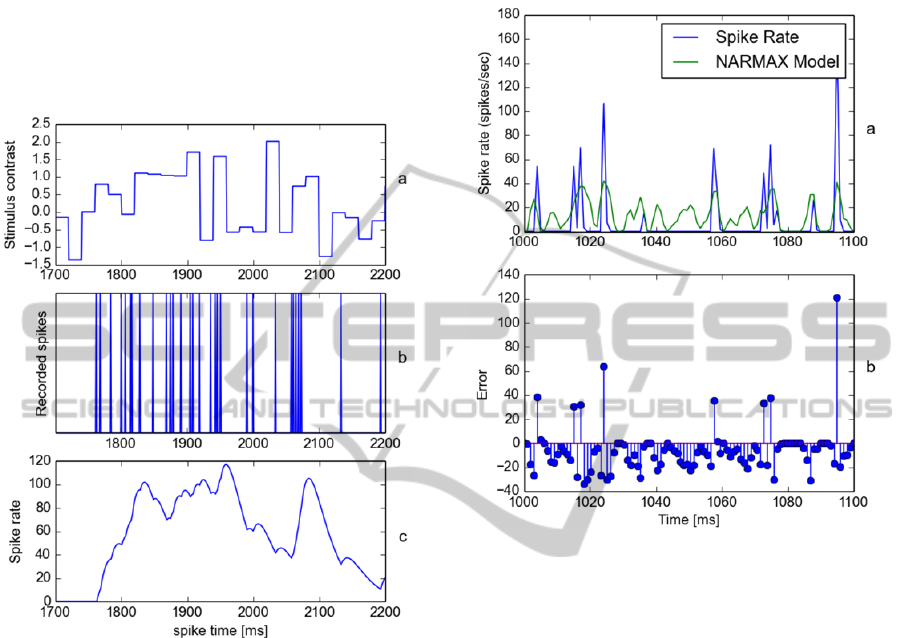
illustrates the spike computed using a half wave
rectified α function.
After this pre-processing stage we can use the
system identification method to obtain a polynomial
model that models the ganglion cells spike rate as a
function of the stimulus intensity where the spatially
uniform stimulus intensity is used as input (Figure
2(a)) and the computed spike rate (Figure 2(c)) is
used as output.
Figure 2: (a) Temporal stimulus intensity; (b) recorded
spikes; (c) computed spike rate for 500ms of a recording.
4.2 Identifying the RGC Linear Model
Using the NARMAX procedure outlined in Section
3 we construct a NARMAX model with an input
regression of nu = 10, corresponding to 200ms of
stimulus time and a polynomial of order 1. The
resulting model contained 11 terms and is presented
in equation (2).
nr(t)= +10.8820504569
+0.4133355718 * u1(n-1)
+5.7712836251 * u1(n-2)
+11.1532223508 * u1(n-3)
+5.4095799493 * u1(n-4)
-0.9100060568 * u1(n-5)
-2.2967796022 * u1(n-6)
-1.5449416639 * u1(n-7)
-0.8126715957 * u1(n-8)
-1.1259153820 * u1(n-9)
-1.4256504406 * u1(n-10)
(2)
Using a new test stimulus sequence we then evaluate
the performance of this NARMAX model and
compare it to the actual neuronal response to the test
stimulus. Results are presented in Figure 3.
Figure 3: Comparison of linear polynomial NARMAX
model and actual neuron response to novel test stimulus
sequence.
Figure 3 (a) illustrates the actual recorded spike rate
(blue) and the linear NARMAX model predicted
spike rate (green). Figure 3 (b) illustrates the model
error. We have also computed the RMSE as 20.92
and present a full comparison of RMSE results in
Table 1.
4.3 Identifying a RGC Quadratic
Model
Again using the same NARMAX procedure we
construct a quadratic NARMAX model with an
input regression of nu = 10, corresponding to 200ms
of stimulus time and a polynomial of order 2. The
resulting quadratic polynomial model contained 26
terms and is presented in equation (3).
Figure 4 (a) illustrates the actual recorded
spike rate (blue) and the quadratic NARMAX model
predicted spike rate (green). Figure 4 (b) illustrates
the model error. We have also computed the RMSE
as 18.31 and present a full comparison of RMSE
results in Table 1. Visual examination reveals both
ModellingandAnalysisofRetinalGanglionCellsThroughSystemIdentification
161
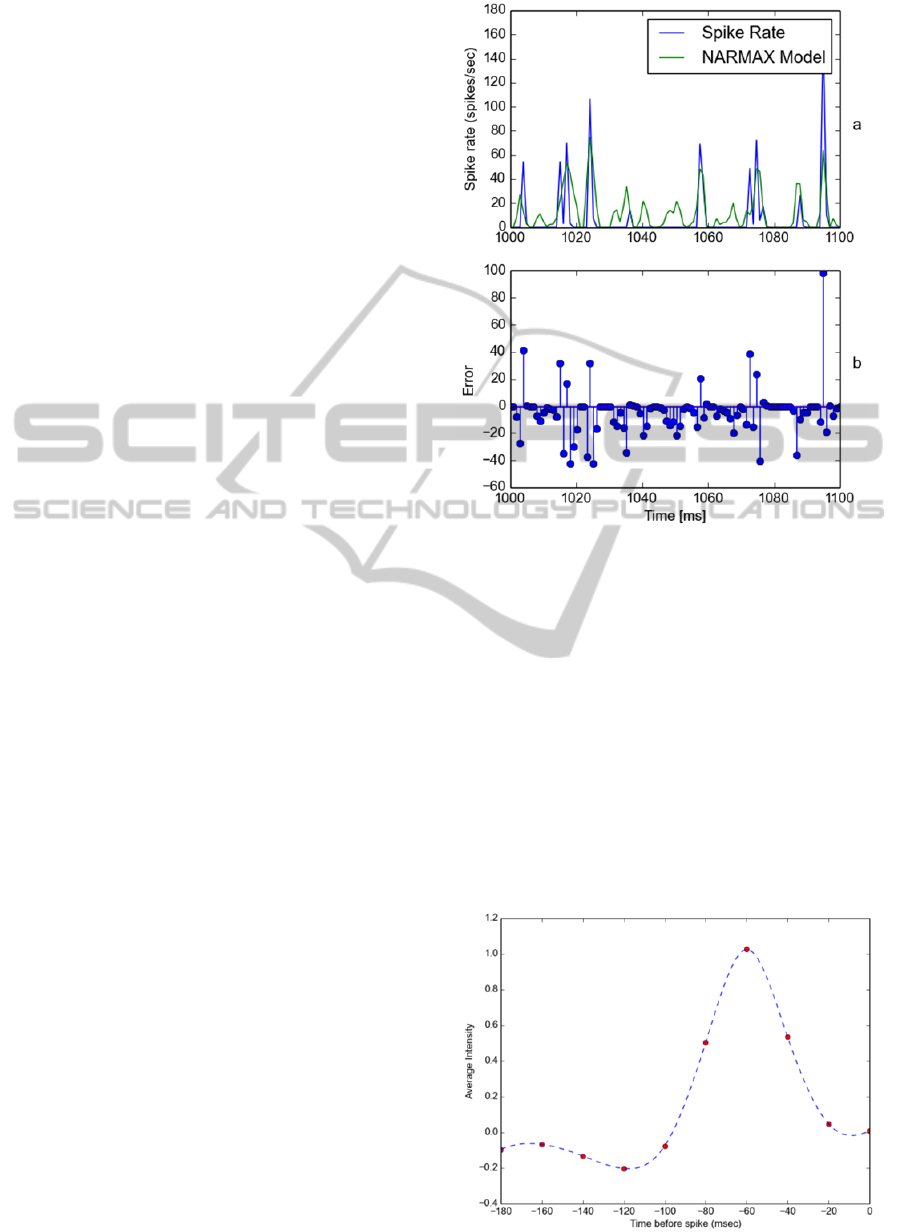
the linear and quadratic NARMAX models perform
similarly with similar errors although the quadratic
model has a reduced error compared to the linear
model.
nr(t)= +5.96244435
+5.70629880 * u1(n-2)
+11.10949549 * u1(n-3)
+5.38644092 * u1(n-4)
-0.89662644 * u1(n-5)
-2.17791043 * u1(n-6)
-1.41390802 * u1(n-7)
-0.75777998 * u1(n-8)
-1.08523454 * u1(n-9)
-1.33837116 * u1(n-10)
+1.65057833 * u1(n-2)^2
+4.28603998 * u1(n-3)^2
-1.01260994 * u1(n-5)^2
+4.84809445 * u1(n-2) * u1(n-3)
+1.55113103 * u1(n-2) * u1(n-4)
-0.63963641 * u1(n-2) * u1(n-5)
-0.71240927 * u1(n-2) * u1(n-6)
+3.30083736 * u1(n-3) * u1(n-4)
-1.14246640 * u1(n-3) * u1(n-5)
-1.76540112 * u1(n-3) * u1(n-6)
-0.95766543 * u1(n-3) * u1(n-7)
-0.66097265 * u1(n-3) * u1(n-9)
-1.10149068 * u1(n-3) * u1(n-10)
-2.13889232 * u1(n-4) * u1(n-5)
-1.68797033 * u1(n-4) * u1(n-6)
-1.03141419 * u1(n-5) * u1(n-6)
(3)
4.4 Comparison with Linear-nonlinear
Model
To provide further comparison for the NARMAX
models we evaluate against a standard benchmark by
computing the Linear-Nonlinear (LNL) model
(Ostojic, 2011). The first stage in computing the
Linear-Nonlinear model is to compute the spike
triggered average (STA) which is the average
stimulus preceding a spike.
To compute the STA, the stimulus in the time
window preceding each spike is extracted, and the
resulting (spike-triggered) stimuli are averaged.
Using the same dataset as the previous analysis we
compute the STA and the results are presented in
Figure 5. Here we can see that the RGC is an ON
cell due to the positive peak in the temporal response
of the filter. We can also see that the cell has a
temporal memory of approximately 150-100ms. The
plot illustrates the average values of stimulus
intensity that elicit a response from the cell.
Figure 4: Comparison of quadratic polynomial NARMAX
model and actual neuron response to novel test stimulus
sequence.
The second stage in the Linear-Nonlinear
model is used to re-construct the ganglion cells
nonlinearity. We using the standard approach
(Ostojic, 2011) of plotting the actual response
against the STA predicted response, binning the
values and fitting a curve using a cumulative density
function. Figure 6 illustrates the obtained non-
linearity. Next, we apply the STA and nonlinearity
to the same test stimulus and compute the response.
We compare this estimated response to the actual
neuronal response and plot the results as before.
Results are presented in Figure 7.
Figure 5: Computed spike triggered average for ON cell.
NCTA2014-InternationalConferenceonNeuralComputationTheoryandApplications
162
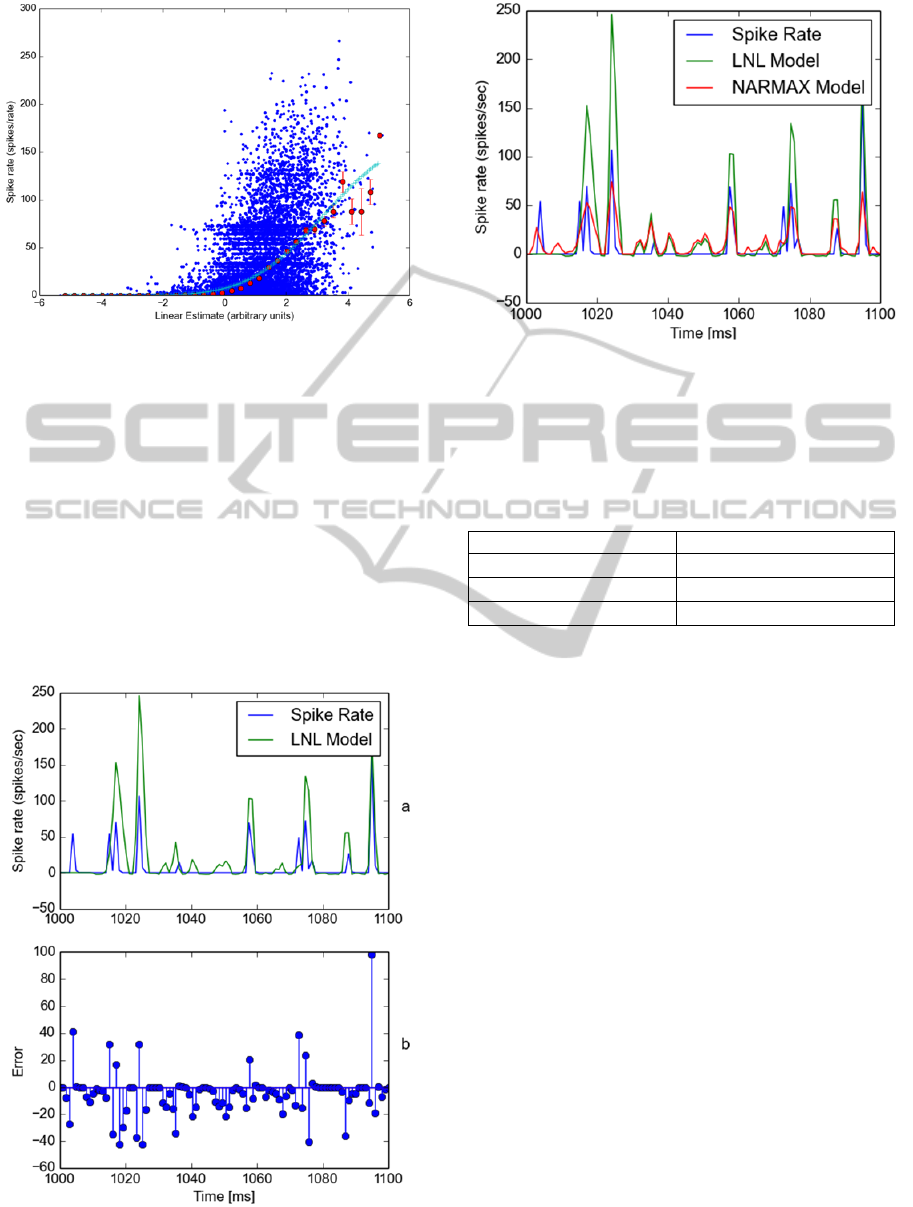
Figure 6: Nonlinearity computed for ON cell.
Figure 7 (a) illustrates the actual recorded spike rate
(blue) and the LNL model predicted spike rate
(green). Figure 7 (b) illustrates the model error. We
have also computed the RMSE as 32.29 and present
a full comparison of RMSE results in Table 1. The
RMSE results illustrate that the quadratic NARMAX
model performs best. We have also plotted the actual
neural response, LNL model predicted response and
the quadratic NARMAX model predicted response
in Figure 8 to illustrate comparative model accuracy.
Visual examination reveals the quadratic NARMAX
model has improved accuracy over the LNL model
and is comparable to the actual neural response.
Figure 7: Comparison of Linear-Nonlinear model and
actual neuron response to novel test stimulus sequence.
Figure 8: Comparative evaluation of actual spike rate,
linear nonlinear model and quadratic NARMAX model.
Table 1: Summary of RMSE for linear NARMAX,
quadratic NARMAX and Linear-Nonlinear models. The
computed RMSE values illustrate that the quadratic
NARMAX approach results in the best fitting model for
the selected dataset.
Method RMSE
Linear NARMAX
20.92
Quadratic NARMAX
18.31
Linear-Nonlinear model
32.29
5 CONCLUSIONS
Modelling biological systems is difficult due to
insufficient knowledge about the internal
components and organisation, and the complexity of
the interactions within the system. Existing
computational models of visual neurons can be
derived by quantitatively fitting particular sets of
physiological data using an input-output analysis
where a known input is given to the system and its
output is recorded as illustrated in the Linear-
Nonlinear approach in Section 4.4.
At a computational level we have presented
the use of the NARMAX system identification
technique to accurately model individual retinal
ganglion cells as shown in Section 4.2 and Section
4.3. We have presented a comparison of the actual
neuronal response and provided a comparison with
the actual neuronal response. Visual comparison
illustrates that all the methods can model the
neuronal response with accuracy although some
errors are present. Computing the RMSE provides a
more quantitative measure of error and the results
summarised in Table 1 illustrate that the quadratic
NARMAX model performs substantially better than
ModellingandAnalysisofRetinalGanglionCellsThroughSystemIdentification
163

both the LNL model and the linear NARMAX
model.
Using NARMAX system identification
techniques to express the biological input-output
coupling mathematically we have modelled highly
complex neuronal structures, and thus "identified"
ganglion cell behaviour with visual scenes. These
polynomial models represent the mapping between
perception and response. The next stage in this work
will be to increase the complexity of the stimulus by
having spatially varying stimuli; we have already
started to test the effectiveness of this using the
natural image sequences.
ACKNOWLEDGEMENTS
The research leading to these results has received
funding from the European Union Seventh
Framework Programme (FP7-ICT-2011.9.11) under
grant number [600954] (“VISUALISE"). The
experimental data contributing to this study have
been supplied by the “Sensory Processing in the
Retina" research group at the Department of
Ophthalmology, University of Göttingen as part of
the VISUALISE project.
REFERENCES
Herikstad, R., Baker, J., Lachaux, J.-P., Gray, C. M., &
Yen, S.-C. (2011). Natural Movies Evoke Spike Trains
with Low Spike Time Variability in Cat Primary
Visual Cortex. Journal of Neuroscience, 31(44),
15844-15860. doi:10.1523/JNEUROSCI.5153-10.
2011
De Boer, Kuyper, P. (1968). “Triggered Correlation”.
Biomedical ngineering, vol.BME-15, no.3, pp.169-
179. doi: 10.1109/TBME.1968.4502561 Transactions
Sakai,H.M., Naka K.I., Korenberg, M.J. (1988) "White-
noise analysis in visual neuroscience". Visual
Neuroscience, 1, pp 287-296 DOI: 10.1017
Chichilnisky EJ (2001) A simple white noise analysis of
neuronal light responses. Network 12(2):199-213.
Talebi, V., Baker, C.L. (2012). "Natural versus Synthetic
Stimuli for Estimating Receptive Field Models: A
Comparison of Predictive Robustness". The Journal of
Neuroscience, Vol. 32, No. 5., pp. 1560-1576,
doi:10.1523
Marmarelis, P.Z., Naka, K.I. (1972). White-noise analysis
of a neuron chain: An application of the wiener theory.
Science 175, 1276–1278
Victor, J., Shapley, R., Knight, B. (1977). Nonlinear
analysis of cat retinal ganglion cells in the frequency
domain. Proc. Natl. Acad. Sci. U.S.A. 74(7), 3068–
3072
Victor, J. (1979) Nonlinear systems analysis: comparison
of white noise and sum of sinusoids in a biological
system. Proc. Natl. Acad. Sci. U.S.A. 76(2), 996–998
Marmarelis, V. (2004) Nonlinear Dynamic Modeling of
Physiological Systems. Wiley Interscience, Hoboken.
Korenberg, M., Hunter, I. (1996). The identification of
nonlinear biological systems: Volterra kernel
approaches. Ann. Biomed. Eng. 24(2), 250–268.
Marmarelis VZ, Zhao X. (1997). Volterra models and
three-layer perceptions. IEEE Trans Neural Networks
8:1421.
Block-oriented Nonlinear System Identification (2010),
Lecture Notes in Control and Information Sciences,
Springer Berlin / Heidelberg, Vol. 404. Giri, F. and
Bai E.W. Eds
Nehmzow, U. (2006) Scientific Methods in Mobile
Robotics: quantitative analysis of agent behaviour.
Springer, 2006.
Ostojic S, Brunel N (2011) From Spiking Neuron Models
to Linear-Nonlinear Models. PLoS Comput Biol 7(1):
e1001056. doi:10.1371/journal.pcbi.1001056
Pillow JW, Paninski L, Uzzell VJ, Simoncelli EP,
Chichilnisky EJ. (2005). Prediction and decoding of
retinal ganglion cell responses with a probabilistic
spiking model. J Neurosci.23;25(47):11003-13.
Korenberg MJ. (1991). Parallel cascade identification and
kernel estimation for nonlinear systems. Ann Biomed
Eng 19:429.
Pillow JW, Shlens J, Paninski L, Sher A, Litke AM,
Chichilnisky EJ, Simoncelli EP. (2008) Spatio
temporal correlations and visual signaling in a
complete neuronal population. Nature 454: 995-999
Billings SA, Voon WSF. 1984. Least-squares parameter
estimation algorithms for non-linear systems. Int.J
Systems Sci 15:601.
Friederich, U., Coca, D., Billings, S.A., Juusola, M. (2009)
Data Modelling for Analysis of Adaptive Changes in
Fly Photoreceptors. Proceedings of the 16th
International Conference on Neural Information
Processing: Part I
Z. Song, S.A. Billings, D. Coca, M. Postma, R.C. Hardie,
& M. Juusola. (2009), Biophysical Modeling of a
Drosophila Photoreceptor. LNCS (ICONIP 2009, Part
I) 5863: 57–71.
Kerr, D, Nehmzow, U and Billings, S.A. (2010) Towards
Automated Code Generation for Autonomous Mobile
Robots. In: The Third Conference on Artificial
General Intelligence, Lugano. Switzerland. Atlantis
Press, Scientific Publishing, Paris, France. 5 pp.
Billings, S.A. Nonlinear system identification: NARMAX
methods in the time, frequency, and spatio-temporal
domains. John Wiley & Sons, 2013.
Lau B, Stanley GB, Dan Y (2002) Computational subunits
of visual cortical neurons revealed by artificial neural
networks. Proc Natl Acad Sci U S A 99: 8974–8979.
Prenger, R., Wu, M.C.K., David, S.V., Gallant, J.L.,
(2004) Nonlinear V1 responses to natural scenes
revealed by neural network analysis, Neural Networks,
Volume 17, Issues 5–6, Pages 663-679, 10.1016/
j.neunet.2004.03.008.
NCTA2014-InternationalConferenceonNeuralComputationTheoryandApplications
164
