
Applications of Genetic Algorithm on Optimal Sequence
for Parrondo Games
Degang Wu and Kwok Yip Szeto
Department of Physics, The Hong Kong University of Science and Technology, Clear Water Bay, Hong Kong, Hong Kong
Keywords:
Genetic Algorithm, Parrondo Game, Optimization, Game Theory.
Abstract:
Parrondo game, which introduction is inspired by the flashing Brownian ratchet, presents an apparently para-
doxical situation where there are ways to combine two losing games into a winning one. The original Parrondo
game consists of two individual games, game A and game B. Game A is a slightly losing coin-tossing game.
Game B has two coins, with an integer parameter M. If the current cumulative capital (in discrete unit) is a
multiple of M, an unfavorable coin p
b
is used, otherwise a favorable p
g
coin is used. Game B is also a losing
game if played alone. Paradoxically, combination of game A and game B could lead to a winning game,
either through random mixture, or deterministic switching. In deterministic switching, one plays according
to a sequence such as ABABB. Exhaustive search and backward induction have been applied to the search
for optimal finite game sequence. In this paper, we apply genetic algorithm (GA) to search for optimal game
sequences with a given length N for large N. Based on results obtained through a problem-independent GA,
we adapt the point mutation operator and one-point crossover operator to exploit the structure of the optimal
game sequences. We show by numerical results the adapted problem-dependent GA has great improvement in
performance.
1 INTRODUCTION
Inspired by the flashing ratchet, Parrondo invented the
games of chance later known as Parrondo game (Par-
rondo, 1996), in which two losing games can be com-
bined following a random or periodic strategy leading
to a winning game. Later, (Allison and Abbott, 2002)
and (Toral et al., 2003) demonstrated that Parrondo
game can be described by a discrete Fokker-Planck
equation, thus a more rigorous relation between Par-
rondo game and Brownian ratchet was established.
From the perspective of game, the optimal sequence
for a given set of parameters for Parrondo games was
inferred by Dinis using dynamic programming (Di-
nis, 2008). It is worth noting that greedy algorithms
do not work in finding the optimal game sequence for
a finite length.
Multi-player versions of the games have been pro-
posed, and they exhibit counterintuitive phenomena
resembling those observed in game, control, and op-
timization theories or economics. Many researchers
have found that greedy algorithms or strategies may
lead to suboptimal or even losing solutions in these
models (Dinis and Parrondo, 2003; Dins and Par-
rondo, 2004; Parrondo et al., 2007). Optimiza-
tion problems of this type warrant the use of meta-
heuristic algorithm such as genetic algorithms (Gold-
berg and Holland, 1988; Holland, 1975).
The successful application of genetic algorithm
has been demonstrated in the solution of many en-
gineering problems, such as speed control of brush-
less DC motor (Xia et al., 2004), cyclic-steam oil
production optimization problem (Patel et al., 2005),
traveling salesman problem (Jiang et al., 2000), mo-
bile robot motion control (Messom, 2002), model-
ing adaptive agents in stock markets (Fong and Szeto,
2001; Szeto and Fong, 2000), and the problem of air-
port scheduling (Shiu and Szeto, 2008). In science,
genetic algorithm has also been used with good re-
sults in fields such as biology (Clote, 2005; Ding and
Zhang, 2008; Pond et al., 2006), clusters (Doye, 2002;
Wales et al., 2000; Wales et al., 1998) and glass tran-
sition (Debenedetti and Stillinger, 2001) in condensed
matter physics.
Our objective of this paper is to use genetic algo-
rithm on the search for optimal game sequence of long
length. We first use Dinis algorithm to gain insight
of the structure of optimal sequences with moderate
length. We then propose structure-preserving evo-
lution operators (mutation and one-point crossover
30
Wu D. and Szeto K..
Applications of Genetic Algorithm on Optimal Sequence for Parrondo Games.
DOI: 10.5220/0005070400300037
In Proceedings of the International Conference on Evolutionary Computation Theory and Applications (ECTA-2014), pages 30-37
ISBN: 978-989-758-052-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

operators), coupled with an appropriate structure-
preserving population initialization procedure to find
the optimal sequence for longer game sequence. Here
structure-preserving condition means that the search
process impose certain constraints on the patterns that
are observed in the optimal sequences of moderate
length, as found either by exhaustive search or Di-
nis algorithm. In order to assess the effect of the
constraints on the quality of the solution, we first
use simple genetic algorithm as a biased sampling
method in order to justify the adaptive approach we
use later. Our paper is organized as follows. In Sec-
tion 2, we introduce the Parrondo game and discuss
the Dinis algorithm that reveals some specific patterns
which appear in the optimal sequence of moderate
length. In Section 3, we propose structure-preserving
mutation and one-point crossover and corresponding
structure-preserving population initialization proce-
dure. In Sections 4 and 5, we discuss the experiment
design and present the experiment results. Finally, the
concluding remarks can be found in Section 6.
2 PARRONDO GAMES
The original Parrondo game consists of two individ-
ual coin tossing games, namely game A and game B.
Game A has only one coin, whose winning probabil-
ity is p
A
= 1/2 − ε, where ε is a small and positive
number. Let X(t) be the cumulative capital of the
player at time t, an integer. If the player keeps playing
game A, the average capital satisfies
hX(t + 1)i = hX(t)i + 2p
A
+ 1 (1)
where h·i is understood as ensemble average. Game
B has two coins, one “good” coin and one “bad” coin.
Game B has an integer parameter M. If X(t) is a
multiple of M, then X(t + 1) is determined by the
“bad” coin with winning probability p
b
= 1/10 − ε,
otherwise the “good” coin with winning probability
p
g
= 3/4 − ε is used. Similar to game A, if the player
keeps playing game B only, the average capital satis-
fies
hX(t + 1)i = hX(t)i + 2[π
0
(t)p
b
+ (1 − π
0
(t))p
g
] − 1
(2)
which explicitly depends on π
0
(t), the probability that
the capital X(t) = 0 mod M. (Harmer and Abbott,
1999) showed that game B is a losing game when
p
b
= 1/10 −ε, p
g
= 3/4 −ε and M = 3, with positive
ε. In this paper, we only discuss the case when M = 3.
There is a recent article discussing the phase diagram
of the more complex situation where the games are
two B games with different M (Wu and Szeto, 2014).
Such extended Parrondo game with multiple M ex-
hibits interesting phenomena such as weak and strong
Parrondo effects when the sequence is infinite.
If we model the Parrondo game as a discrete-
time Markov chain as in (Harmer and Abbott,
2002), we can define the probability vector π
π
π(t) ≡
(π
0
(t), π
1
(t), π
2
(t))
T
(for M = 3). Accordingly, the
transition matrix for game A is
Π
A
=
0 1 − p
A
p
A
p
A
0 1 − p
A
1 − p
A
p
A
0
(3)
such that the time evolution equation is π
π
π(t + 1) =
Π
A
π
π
π(t). Similarly, the transition matrix for game B is
Π
B
=
0 1 − p
g
p
g
p
b
0 1 − p
g
1 − p
b
p
g
0
(4)
Parrondo game can be played according to a deter-
ministic finite game sequence such as ABABB, so at
the end of the sequence, the probability vector π
π
π(5) is
π
π
π(5) = Π
B
Π
B
Π
A
Π
B
Π
A
π
π
π(0). (5)
Algorithm 1: Function to calculate the expected return
for game sequence {S
i
} with length N.
function EXPECTED RETURN({S
i
})
g ← 0
π
π
π = [1,0,0]
T
for t ← 1,N do
if S
t
= A then
g ← g + π
π
π
T
[2p
A
− 1,2p
A
− 1,2p
A
− 1]
T
π
π
π ← Π
A
π
π
π
else
g ← g + π
π
π
T
[2p
b
− 1,2p
g
− 1,2p
g
− 1]
T
π ← Π
B
π
π
π
end if
end for
return g
end function
Parrondo game has a seemingly paradoxical prop-
erty that while game A and B are losing when they are
played individually, the stochastic mixture of game
A and B, or playing according to a deterministic se-
quence may lead to a winning combined game for
small positive value of ε. For the detailed analysis
of this paradox, please refer to (Harmer and Abbott,
2002). For a finite game sequence with length N,
the expected return at the end of the game sequence
can be computed by Algorithm 1. Our task is to find
the finite game sequence that has maximum cumula-
tive gain. The expected return per game in the sta-
tionary state for a periodic sequence with length N
ApplicationsofGeneticAlgorithmonOptimalSequenceforParrondoGames
31

can be computed using Algorithm 1 with two mi-
nor modifications: the initial value of π
π
π should be
the equilibrium distribution of the transition matrix
Π
α(1)
Π
α(2)
·· ·Π
α(N)
and the final value of g should
be divided by N.
Sequences up to period 12 have been studied
using symbolic manipulators and exhaustive search
(Wagon and Velleman, 2001), and the periodic se-
quence ABABB, or any of its permutations, has come
up as the best in the sense that it provides the high-
est returns in the stationary state. However, exhaus-
tive search for optimal sequences of finite length N
is not feasible for large N. Using dynamic program-
ming, Dinis discovered that optimal sequences with
finite N “consist of several repetitions of the ABABB
motif flanked by brief pieces of other sequences. (Di-
nis, 2008) For example, the optimal sequence for
N = 20 with initial condition X (t) = 0 and ε = 0 is
AB ABABBABABBABABB ABB. In fact, the struc-
ture of the optimal sequences is more specific: they
are strings of AB and ABB. From these results, we
have the following definition:
Definition 1. A game sequences is said to have a spe-
cial structural-property if it is made of AB and/or ABB
substrings exclusively.
Straightforward implementation of Dinis algo-
rithm requires storage space that scales as N, in or-
der to store all the numerical results from all inter-
mediate steps. Moreover, the algorithm approximates
two-dimensional plane with a discrete square grid (In
(Dinis, 2008), a 2000 × 2000 grid was used for each
step). The error in the expected payoff due to grid ap-
proximation will accumulate across time and the total
error in the expected payoff at the end of the sequence
is at least a linear function of N. The effect of this
error to the correctness of the optimal sequence has
not been investigated. The approximation therefore
constraints the computation of optimal sequences for
large N. This suggests that heuristic algorithms such
as GA that demands much less space and does not re-
quire approximation in the expected payoff are useful
for optimal sequences with long length. Moreover,
by definition, Dinis’ algorithm does not provide any
information of the suboptimal sequence. In contrast,
while searching for the optimal solution, GA also ef-
ficiently samples many suboptimal solutions, which
provide insights in the relation between the structure
of a sequence and its performance.
3 STRUCTURE-PRESERVING
GENETIC ALGORITHM
3.1 Representation
If we map game A to 0 and map game B to 1, a game
sequence with length N can be mapped to a binary
string with the same length, and the order of the bi-
nary values corresponds to the order the games are
played. The objective function is given by Algorithm
1. For example, the game sequence ABABB is en-
coded as 01011. For game sequences that are strings
of AB and ABB, an auxiliary representation is use-
ful: 0 for AB and 1 for ABB. For example, game
sequence ABABB is encoded as 01 in this auxiliary
representation. However, game sequences under this
auxiliary representation vary in length from sequence
to sequence. To distinguish these two kinds of repre-
sentations, we call the one that maps game A to 0 the
original representation.
3.2 Simple Genetic Algorithm
A simple genetic algorithm is used as the reference.
Algorithm 2: “Simple” Genetic Algorithm.
procedure SGA
INIT(P) P for the population
repeat
P
0
← DET TOURNAMENT(P,k)
for i = 0,SIZE(P)/2-1 do
if RAND()< p
c
then
XOVER(P
0
(2i),P
0
(2i + 1))
end if
end for
for i = 0,SIZE(P)-1 do
for j = 0, N − 1 do
if RAND()< p
m
then
FLIP(P
0
(i, j))
end if
end for
end for
P ← P
0
EVALUATE(P) update the fitness values
until Stopping criteria satisfied
end procedure
In the initialization, each locus of each chromo-
some is set to 1 or 0 with equal probability. The new
population P
0
is generated from the last one using
deterministic tournament: from previous generation,
randomly draw k chromosomes with equal probabil-
ity, and select the one with the highest fitness value.
ECTA2014-InternationalConferenceonEvolutionaryComputationTheoryandApplications
32

This process is repeated until the new population has
the same number of chromosomes as the previous
population. The one-point crossover operator is de-
noted by XOVER(), and its rate is denoted by p
c
. The
mutation rate per bit is denoted by p
m
.
3.3 Structure-preserving Mutation
Operator
If a chromosome has special structural property, we
can apply a mutation operator that preserves this prop-
erty. We propose a structure-preserving mutation op-
erator in Algorithm 3.
Algorithm 3: Structure-preserving Mutation Operator.
function SPMUTATE(c) c for a chromosome
c
0
← ORIGINTOAUX(c)
for j = 0,SIZE(c
0
)−1 do
if RAND()< p
m
then
FLIP(c
0
( j)) c
0
( j) is the j-th locus of c
0
c ←AUXTOORIGINAL(c
0
)
end if
end for
return c
end function
However, when we use Algorithm 3, we must
check if the sequence after mutation has length N.
We therefore need an algorithm for chromosome re-
pair, using first a conversion function AUXTOORIG-
INAL(c
0
). This function converts c
0
in the auxiliary
representation back to a chromosome in the original
representation, which is not always possible because
the converted chromosome in original form may have
a length greater or less than N. If SIZE(c)> N, we then
pop 01 or 011 from the back of c until SIZE(c)< N.
We append 01 or 011 to the back of c such that
SIZE(c)= N.
3.4 Structure-preserving Crossover
Operator
If a chromosome has special structural property, we
can apply an adapted one-point crossover operator
that preserves this propose. The structure-preserving
one-point crossover operator is shown in Algorithm 4.
Here the function CHOOSEXPT decides the
crossover point in the auxiliary representation such
that the two children chromosomes in the original rep-
resentation are either one bit longer or shorter than N.
The function FIXCHROMOSOME lengthens or short-
ens the chromosome by mutating 01 to 011, 011 to 01,
converting 011 to 0101, or converting 0101 to 011.
Algorithm 4: Structure-preserving One-point cross-
over operator.
procedure SPXOVER(c
1
,c
2
)
c
0
1
← AUXREPRE(c
1
)
c
0
2
← AUXREPRE(c
2
)
xPt ← CHOOSEXPT(c
0
1
,c
0
2
)
XOVER(c
0
1
,c
0
2
,xPt)
c
0
1
← FIXCHROMOSOME(c
0
1
)
c
0
2
← FIXCHROMOSOME(c
0
2
)
c
1
← AUXTOORIGINAL(c
0
1
)
c
2
← AUXTOORIGINAL(c
0
2
)
end procedure
3.5 Structure-preserving Genetic
Algorithm
In order to replace the mutation operator and one-
point crossover operator with the structure-preserving
versions, the chromosomes must have special struc-
tural property at all time. For this purpose, a structure-
preserving initialization routine is propose in Algo-
rithm 5.
As we will show later, this initialization routine
per se greatly improves the quality of the initial chro-
mosomes. In this paper, the structure-preserving ge-
netic algorithm (SPGA) refers to ones that run in the
framework of Algorithm 2, with population initial-
ized by Algorithm 5, the mutation operator replaced
by Algorithm 3 and the one-point crossover opera-
tor replaced by Algorithm 4. This combination of
algorithms in SPGA reduces the search space sub-
stantially. One concern about this reduction is that
it might fragment the solution space into disjoint sub-
space. We will discuss this issue in Section 5.
Algorithm 5: Structure-Preserving Initialization.
procedure SPINIT(P)
for each c
i
in P do
empty c
i
append 01 or 011 randomly to c until
SIZE(c)==N
end for
end procedure
4 EXPERIMENTAL DESIGN
For comparison of the relative efficiency of the vari-
ous components of our genetic algorithm, we consider
the following versions of GAs.
ApplicationsofGeneticAlgorithmonOptimalSequenceforParrondoGames
33
• SGANoX: A GA that uses only the mutation oper-
ator, with uniform initialization and no crossover
operators
• SGANoXwSPInit: SGANoX with structure-
preserving initialization
• SGA: Simple Genetic Algorithm, with uniform
initialization, standard mutation operator and one-
point crossover operator
• SGAwSPInit: Simple Genetic Algorithm with
structure-preserving initialization, standard muta-
tion operator and one-point crossover operator
• SPGANoX: Structure-preserving mutation only
genetic algorithm with structure-preserving ini-
tialization and structure-preserving mutation op-
erator, but without crossover
• SPGA: Structure-preserving genetic algorithm
with structure-preserving initialization, structure-
preserving mutation operator and structure-
preserving crossover operator
For simplicity, when we use the term SGAs, we
mean the group of SGA, including SGANoX, SGA,
SGANoXwSPInit, SGAwSPInit. Similarly, when we
use the term SPGAs, we mean the group of SPGA,
including SPGANoX, SPGA.
The size of the population, N
P
, will be set to
100 for all versions of GAs. The size of the tourna-
ment, k, defined in Algorithm 2 in the deterministic
tournament selection process will be set to 10. The
crossover rate, p
c
and the mutation rate per bit, p
m
,
both defined in Algorithm 2, will be chosen individu-
ally for each GA such that each GA has good perfor-
mance.
5 RESULTS AND ANALYSIS
First we investigate the effect of imposing structure-
preserving evolution operators and population initial-
ization on search space. Exhaustive search is not an
option for sequences, as long as N = 80. Instead
we use SGA to perform a biased sampling on the
search space by just running SGA with p
m
= 0.02 and
p
c
= 1.0. The selection mechanism in SGA biases
towards chromosomes having higher fitness values.
Every chromosome that appears during 200 genera-
tions of evolution from 50 independent experiments
are collected as samples. Numerical results show that
the fitness distribution for game sequences satisfy-
ing special structural property are strongly grouped
together at high fitness values, and they are among
the chromosomes having fitness values mostly higher
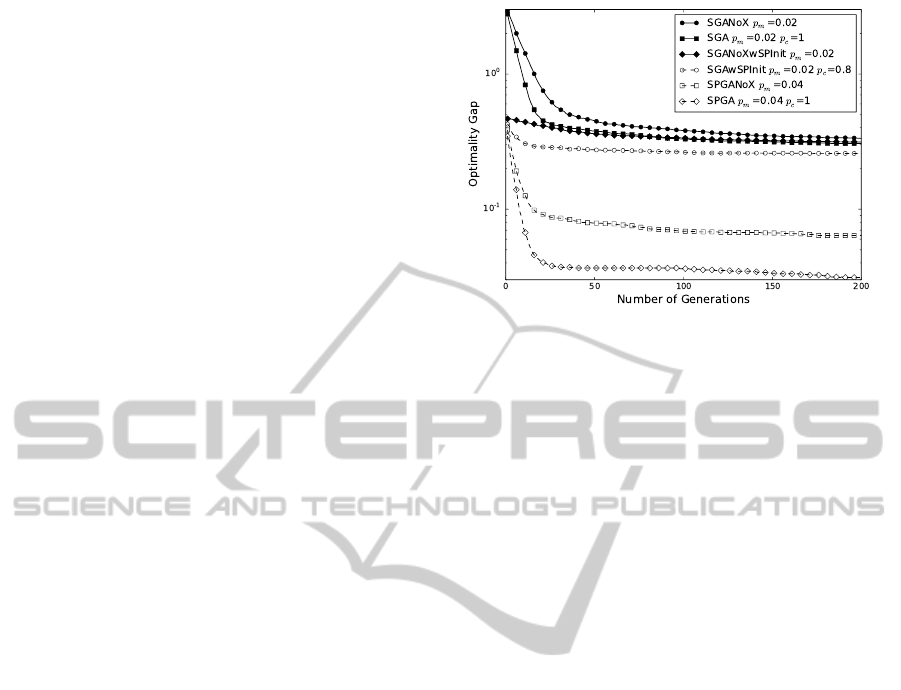
Figure 1: Optimality gap as a function of number of gen-
erations for N = 80. Every curve is an average from 100
independent experiments. Y-axis is in log-scale. The gap is
smallest for SPGA, indicating it is the best performing GA.
than the highest fitness of the sequences without spe-
cial structure. From this observation, we conclude
that imposing structure-preserving evolution opera-
tors and population initialization on search space will
not fragment the search space. Furthermore, the usage
of structure-preserving evolution operators and popu-
lation initialization can be justified due to the overall
higher fitness value obtained.
Next we compare the performances of the various
GAs mentioned in the last section on the search for
optimal sequence of with N = 80. We first calculate
the true expected return of the optimal sequence using
Dinis’ algorithm. We then measure the performance
of various GAs by the optimality gap, the difference
between the expected return of the optimal sequence
and the expected return of the chromosome with the
highest fitness value. We show the optimality gap as
a function of number of generations in Fig. 1. The
values of p
m
and p
c
are chosen on the basis of perfor-
mance.
The structure-preserving initialization alone con-
tributes significantly to the performance, as can be
seen from the relative performances of SGANoX
and SGA with or without structure-preserving pop-
ulation initialization in Fig. 1. In the case
of problem-independent GAs, one-point crossover
operator contributes to the early edge in perfor-
mance. The problem-dependent structure-preserving
GAs generally perform better. SPGANoX outper-
forms SGAwSPInit. SPGA performs the best.
Since there is no guarantee that optimal game se-
quence will be obtained after 200 generations, we
therefore need to assess the average quality of the best
solutions after 200 generations. For N=80, the opti-
mal game sequence is
ABXXXXXXXXXXXXXXXABB, (6)
ECTA2014-InternationalConferenceonEvolutionaryComputationTheoryandApplications
34

where X stands for ABABB. We call those AB (or
ABB) substring that are not able to form ABABB with
adjacent substring ungrouped AB (or ABB).
A typical suboptimal but still good solution re-
sulted from SPGAs has the following structure:
XXXXXXABBXXXXABBXXXABXAB, (7)
which possesses the right number of AB and ABB
substrings, and the SPGAs manage to figure out the
overall structure. From this observation, we expect
that a problem-specific local search can obtain the real
optimal solution from the best solution obtained from
SPGAs. In this context, we may introduce some alter-
native quality measures by the following enquiries:
1. Has the right number of AB and ABB been reached?
(Here the right number refers to the number in the opti-
mal sequence.) If not, calculate the deviation by
d
1
(s) = |N
AB
(s) −N
AB
(s
∗
)| + |N
ABB
(s) − N
ABB
(s
∗
)|,
(8)
where N
AB
(s) (N
ABB
(s)) is the number of AB (ABB re-
spectively) and s
∗
is the optimal sequence.
2. If the right number of AB and ABB has been reached,
has the right number of ungrouped AB and ABB been
reached? (Here, the right number again refers to the
number in the optimal sequence.) If N
∗
AB
(s) (N
∗
ABB
(s))
is the number of ungrouped AB (ABB respectively) and
s
∗
is the optimal sequence, then this deviation can be
written as
d
2
(s) = |N
∗
AB
(s
∗
) − N
∗
AB
(s)| + |N
∗
ABB
(s
∗
) − N
∗
ABB
(s)|.
(9)
For both measures, the smaller the values of d
1
(s)
and d
2
(s) the higher is the quality of the sequence. We
show the statistics of the best chromosomes obtained
by various GAs at the end of 200-th generation in Ta-
ble 1. Data are collected from 100 independent exper-
iments for each GA variant. As can be seen from the
table, SPGAs are better at figuring out the structure of
the optimal sequence than other GAs.
We see that for medium size sequence (N = 80),
our SPGA is good approximation to Dinis analysis, as
shown in Fig. 1. Thus for medium size sequence, we
do not see the advantage of our SPGA algorithm.
We now apply the same set of GAs on searching
for the optimal sequences with longer length N. The
values of p
m
and p
c
are the same as those used be-
fore. Note that the exact optimal sequence with length
N = 200 is not easily available as straightforward im-
plementation of Dinis algorithm needs huge amount
of storage space and exhaustive search will require
even more. Thus, we use an alternative measure of
performance: we record the highest fitness value in
the population as a function of the number of gen-
erations, averaged over 50 independent experiments
and see which GA yields the best result. The perfor-
mances of various GAs as a function of number of
Table 1: Statistics of the best chromosomes obtained by
various GAs at the end of 200-th generation for N = 80.
Data are collected from 100 independent experiments for
each GA variant. The best chromosomes obtained from
SGAs do not necessarily satisfy the special structural prop-
erty, and therefore the numbers of instances of best chro-
mosomes that satisfy the property are indicated. The quan-
tity Prs stands for the Probability that Structural property is
observed over 100 independent experiments. In the calcu-
lation of the quality measure, d
1
(s) and d
2
(s) for the case
of SGA, we only count those chromosomes satisfying the
special structural property.
GA Variant Prs d
1
(s) d
2
(s)
SGANoX 97/100 3.60825 7.87629
SGA 96/100 3.48958 6.84375
SGANoXwSPInit 97/100 4.89691 8.17526
SGAwSPInit 100/100 4.75 6.47
SPGANoX 100/100 0.25 0.89
SPGA 100/100 0.00 0.54
generation when searching for optimal sequence with
length N = 200 are shown in Fig. 2. We see that SP-
GAs outperform problem-independent SGAs. In fact,
for SGAs with structure-preserving initialization, the
average best fitness value drops as the number of gen-
eration increases. Let us compare this curious fea-
ture of decreasing average fitness of SGAs for large
N(= 200) with the results for small N(= 80) shown
in Fig. 1. The optimality gaps are all decreasing with
generation number in Fig. 1, implying that the aver-
age fitness of all GAs are increasing. We also see that
SGAs are able to take advantage of the good qual-
ity initial chromosomes resulted from the structural-
preserving initialization. However, when N is large,
as shown in Fig. 2 for N = 200, SGAs no longer have
the ability in exploiting the good quality of the initial
chromosomes. In Fig. 2, we observe the merging of
several curves at large generation number:
1. SPGA(♦) merges with SPGANoX()
2. SGAwSPInit (◦) merges with SGA()
3. SGANoX(•) merges with SGANoXwSPInit();
We see from 1 that SPGA with crossover operator,
does not have noticeable advantage over SPGANoX
in terms of best fitness value in the population, sug-
gesting structure-preserving crossover operator does
not provide additional advantage for large-size prob-
lem. From 2 we see that for SGA, the special initial-
ization with structure preserving features is irrelevant
for large N. From 3, we see similar effect as 2 without
crossover for SGA, when N is large.
In Table 2, we see the average of the best fit-
ness values at the end of 200-th generation and the
standard deviation. Not only do SPGAs (SPGANoX
and SPGA) achieve higher best fitness value, they
ApplicationsofGeneticAlgorithmonOptimalSequenceforParrondoGames
35
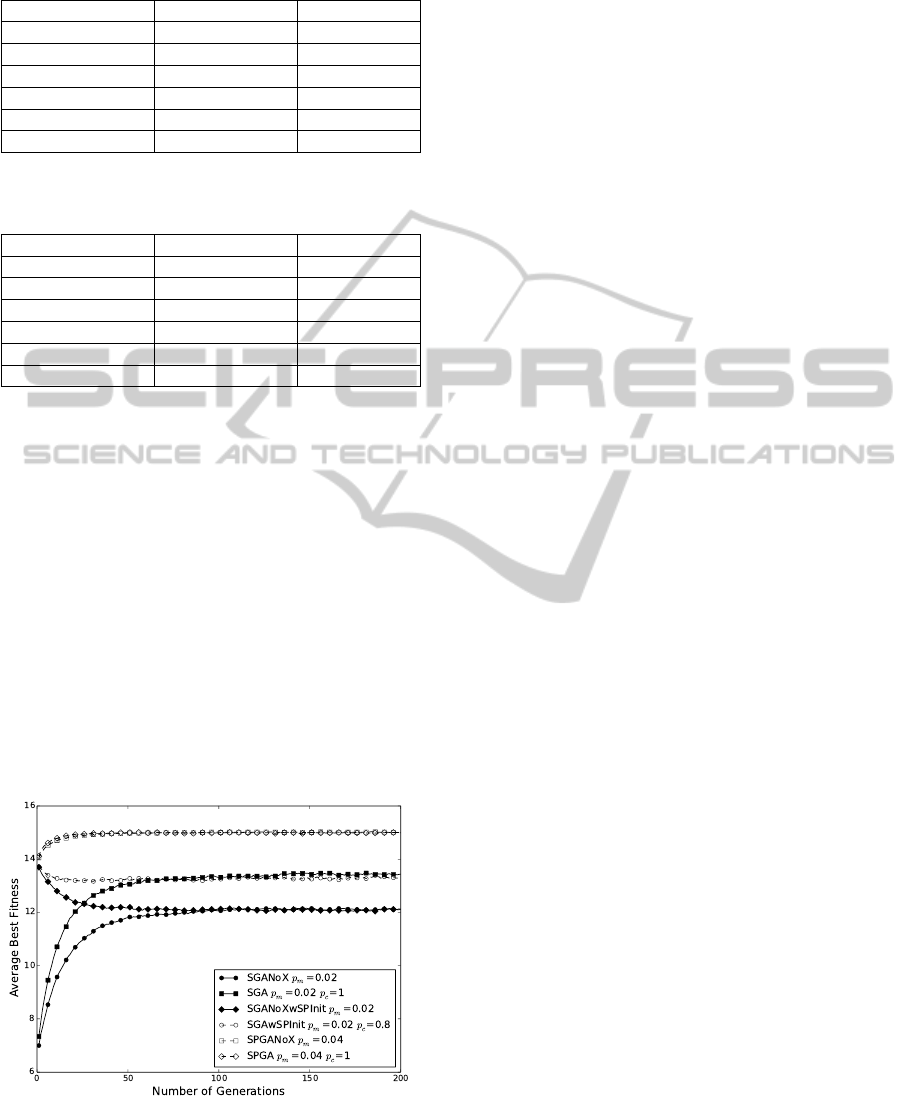
Table 2: Best fitness values statistics for N = 200 at the end
of the experiment. Data are collected from 50 experiments.
GA Variant Avg. best fitness Stand. deviat.
SGANoX 12.1 0.231
SGA 13.4 0.250
SGANoXwSPInit 12.1 0.228
SGAwSPInit 13.3 0.273
SPGANoX 15.01 0.0805
SPGA 15.00 0.0978
Table 3: Best fitness values statistics for N = 300 at the end
of the experiment. Data are collected from 50 experiments.
GA Variant Avg. best fitness Stand. deviat.
SGANoX 15.9 0.364
SGA 18.0 0.360
SGANoXwSPInit 15.9 0.348
SGAwSPInit 17.7 0.334
SPGANoX 21.90 0.0907
SPGA 21.97 0.0893
also have smaller standard deviation in the best fit-
ness value, which means that SPGAs are more re-
liable. Moreover, SPGANoX and SPGA have sim-
ilar performance statistically, suggesting structure-
preserving crossover operator does not provide no-
ticeable additional advantage for large-size problem.
Results for optimal sequence search with length
N = 300 exhibit similar qualitative features (Table
3). SPGAs outperform SGAs in terms of aver-
age best fitness value. SGAs are not able to ex-
ploit the good quality chromosomes resulted from the
structure-preserving initialization. Similar to the case
of N = 200, the structure-preserving crossover op-
erator does not provide additional benefit in perfor-
mance.
Figure 2: Average best fitness as a function of number of
generations for N = 200. Every curve is an average from 50
independent experiments.
6 CONCLUSIONS AND FUTURE
WORK
In this paper, we have proposed problem-dependent
structure-preserving mutation operator, one-point
crossover operator and initialization routine. We have
designed several numerical experiments to evaluate
the relative efficiency of several GAs incorporating
various features deemed important from observation
of small N optimal sequences. We have shown that
the proposed structure-preserving initialization rou-
tine offers high-quality candidate solutions. We have
also shown that both structure-preserving mutation
operator and one-point crossover operator improves
the performance greatly compared to various versions
of simple GAs. The various versions of SPGAs are
consistently better at figuring out the structure of opti-
mal sequence for medium-size problem, e.g. N = 80.
In medium-size problems, e.g. N = 80, structure-
preserving crossover operator provides noticeable ad-
ditional advantage; however, for large-size problems,
e.g. N = 200, structure-preserving crossover oper-
ator does not provide noticeable additional advan-
tage. This observation could be useful in using GAs
to discover the structures of sequences for different
lengths. In general, our analysis using GA for Par-
rondo game sequences can be extended to the analysis
of sequences from other fields. For the future works,
we could develop a permutation-based evolution op-
erator to further improve the performance of SPGAs.
Within the confine of limited numerical analysis
for long sequence, we can conclude that our algorithm
is useful in searching for the optimal sequence for Par-
rondo game. Since the exact solution is not known
for long sequence, our method does provide useful
candidates for benchmarking. This is important for
applications in other sequence analysis for which a
good solution, not the mathematically optimal one,
is needed. In the context of Parrondo game, Dinis
algorithm for short and medium sequences can pro-
vide a benchmark for the optimal solution, which we
can also obtain by SPGA. However, for very long se-
quence, when Dinis algorithm is not easily used, our
SPGA does provide a good solution. In practical ap-
plications in other sequences, we can first analyze the
motif in short sequences where exhaustive search pro-
vide the optimal solution, and then use those motifs to
obtain a good solution for long sequence with SPGA.
ACKNOWLEDGEMENTS
K. Y. Szeto acknowledges the support of grant FS
GRF13SC25and FS GRF14SC28. Degang Wu would
ECTA2014-InternationalConferenceonEvolutionaryComputationTheoryandApplications
36

like to thank Zhangyu Chang for helpful discussions.
REFERENCES
Allison, A. and Abbott, D. (2002). The physical basis
for parrondo’s games. Fluctuation and Noise Letters,
2(04):L327L341.
Clote, P. (2005). An efficient algorithm to compute the land-
scape of locally optimal RNA secondary structures
with respect to the nussinov-jacobson energy model.
Journal of computational biology, 12(1):83101.
Debenedetti, P. G. and Stillinger, F. H. (2001). Su-
percooled liquids and the glass transition. Nature,
410(6825):259267.
Ding, Y.-S. and Zhang, T.-L. (2008). Using chous pseudo
amino acid composition to predict subcellular lo-
calization of apoptosis proteins: An approach with
immune genetic algorithm-based ensemble classifier.
Pattern Recognition Letters, 29(13):1887–1892.
Dinis, L. (2008). Optimal sequence for parrondo games.
Physical Review E, 77(2):021124.
Dinis, L. and Parrondo, J. M. R. (2003). Optimal strate-
gies in collective parrondo games. Europhysics Let-
ters (EPL), 63:319–325.
Dins, L. and Parrondo, J. M. (2004). Inefficiency of voting
in parrondo games. Physica A: Statistical Mechanics
and its Applications, 343:701–711.
Doye, J. P. (2002). Network topology of a potential energy
landscape: A static scale-free network. Physical re-
view letters, 88(23):238701.
Fong, L. Y. and Szeto, K. Y. (2001). Rules extraction in
short memory time series using genetic algorithms.
The European Physical Journal B-Condensed Matter
and Complex Systems, 20(4):569572.
Goldberg, D. E. and Holland, J. H. (1988). Genetic al-
gorithms and machine learning. Machine learning,
3(2):9599.
Harmer, G. P. and Abbott, D. (1999). Game theory: Los-
ing strategies can win by parrondo’s paradox. Nature,
402(6764):864864.
Harmer, G. P. and Abbott, D. (2002). A review of
parrondos paradox. Fluctuation and Noise Letters,
2(2):R71R107.
Holland, J. H. (1975). Adaptation in natural and artificial
systems: An introductory analysis with applications to
biology, control, and artificial intelligence. U Michi-
gan Press.
Jiang, R., Szeto, K. Y., Luo, Y. P., and Hu, D. C. (2000).
Distributed parallel genetic algorithm with path split-
ting scheme for the large traveling salesman problems.
In Proceedings of Conference on Intelligent Informa-
tion Processing, 16th World Computer Congress, page
2125.
Messom, C. (2002). Genetic algorithms for auto-tuning mo-
bile robot motion control.
Parrondo, J. M. R. (1996). How to cheat a bad mathemati-
cian. EEC HC&M Network on Complexity and Chaos.
Parrondo, J. M. R., Dinis, L., Garca-Torao, E., and Sotillo,
B. (2007). Collective decision making and paradox-
ical games. The European Physical Journal Special
Topics, 143(1):39–46.
Patel, A. N., Davis, D., Guthrie, C. F., Tuk, D., Nguyen,
T. T., Williams, J., and others (2005). Optimiz-
ing cyclic steam oil production with genetic algo-
rithms. In SPE Western Regional Meeting. Society of
Petroleum Engineers.
Pond, S. L. K., Posada, D., Gravenor, M. B., Woelk, C. H.,
and Frost, S. D. W. (2006). GARD: a genetic algo-
rithm for recombination detection. Bioinformatics,
22(24):3096–3098.
Shiu, K. L. and Szeto, K. Y. (2008). Self-adaptive mutation
only genetic algorithm: An application on the opti-
mization of airport capacity utilization. In Intelligent
Data Engineering and Automated LearningIDEAL
2008, page 428435. Springer.
Szeto, K. Y. and Fong, L. Y. (2000). How adaptive agents in
stock market perform in the presence of random news:
A genetic algorithm approach. In Intelligent Data
Engineering and Automated LearningIDEAL 2000.
Data Mining, Financial Engineering, and Intelligent
Agents, page 505510. Springer.
Toral, R., Amengual, P., and Mangioni, S. (2003). Par-
rondo’s games as a discrete ratchet. Physica A: Statis-
tical Mechanics and its Applications, 327(1-2):105–
110.
Wagon, S. and Velleman, D. (2001). Parrondo’s para-
dox. Mathematica in Education and Research, 9(3-
4):8590.
Wales, D. J., Doye, J. P., Miller, M. A., Mortenson,
P. N., and Walsh, T. R. (2000). Energy landscapes:
from clusters to biomolecules. Advances in Chemical
Physics, 115:1112.
Wales, D. J., Miller, M. A., and Walsh, T. R.
(1998). Archetypal energy landscapes. Nature,
394(6695):758760.
Wu, D. and Szeto, K. Y. (2014). Extended parrondo’s game
and brownian ratchets: Strong and weak parrondo ef-
fect. Physical Review E, 89(2):022142.
Xia, C., Guo, P., Shi, T., and Wang, M. (2004). Speed con-
trol of brushless DC motor using genetic algorithm
based fuzzy controller. In Proceeding of the 2004
International Conference on Intelligent Mechatronics
and Automation, Chengdu, China, 3rd edn. A Treatise
on Electricity and Magnetism, volume 2, page 6873.
ApplicationsofGeneticAlgorithmonOptimalSequenceforParrondoGames
37
