
Digital Database for Screening Mammography Classification Using
Improved Artificial Immune System Approaches
Rima Daoudi
1,2
, Khalifa Djemal
1
and Abdelkader Benyettou
2
1
IBISC Laboratory, University of Evry Val d’Essonne, 91020 Evry Cédex ,France
2
SIMPA Laboratory, University of Sciences and Technologies, BP 1505 El-M'naouar, Oran, Algeria
Keywords: DDSM, Breast Cancer, Clonal Selection, Local Sets, Median Filter, Clone, Mutate.
Abstract: Breast cancer ranks first in the causes of cancer deaths among women around the world. Early detection and
diagnosis is the key for breast cancer control, and it can increase the success of treatment, save lives and
reduce cost. Mammography is one of the most frequently used diagnosis tools to detect and classify
abnormalities of the breast. In this aim, Digital Database for Screening Mammography (DDSM) is an
invaluable resource for digital mammography research, the purpose of this resource is to provide a large set
of mammograms in a digital format. DDSM has been widely used by researchers to evaluate different
computer-aided algorithms such as neural networks or SVM. The Artificial Immune Systems (AIS) are
adaptive systems inspired by the biological immune system, they are able of learning, memorize and
perform pattern recognition. We propose in this paper several enhancements of CLONALG algorithm, one
of the most popular algorithms in the AIS field, which are applied on DDSM for breast cancer classification
using adapted descriptors. The obtained classification results are 98.31% for CCS-AIS and 97.74% for MF-
AIS against 95.57% for original CLONALG. This proves the effectiveness of the used descriptors in the
two improved techniques.
1 INTRODUCTION
Breast Cancer is malignant tumor that develops from
breast cells; it is becoming one of the major causes
of death among women in the whole world,
according to the World Health Organization (WHO)
the incidence rate of breast cancer between 2008 and
2012 was more than 20% with 14% of mortality rate
(Ferlay and al., 2013), and as there is no prevention
techniques, the only way to help patients survive is
by early detection. If cancerous cells are detected
before they spread other organs, the survival rate of
patient is more than 97% (American cancer society
homepage 2008).
There is no doubt that the evaluations of data
taken from patients and experts decisions are most
important factors in diagnosis. However, artificial
intelligence techniques and expert systems are
gaining popularity in this field by dint of their high
diagnosis capability and effective classification. In
this context, various articles have been published in
the aim of classifying breast cancer databases using
artificial intelligence techniques as Neural Networks
(Marcano-Cedeno et al.,2011)(Timmy Manning and
Paul Walsh, 2013)(Zadehand et al., 2012)(Aboul
Ella Hassanien et al., 2014), SVM(Mahnaz and
Broumandnia, 2013)(Aboul Ella Hassanien et al.,
2013); Genetic Algorithms(Jain, R. and J.
Mazumdar, 2003)(Mazurowski, M.A., et al., 2007)
Expert Systems (Wan Noor et al., 2013) and
Artificial Immune Systems(Sharma and Sharma,
2011)(Daoudi et al., 2013)(Daoudi et al., 2013).
The natural immune system is composed of
diverse sets of cells and molecules that work
together with other systems (like neural and
endocrine) for maintaining homeostatic state. It
primary function is to protect the body from foreign
substances called antigens by recognizing and
eliminating them. This process is known as the
immune response. It makes use of a huge variety of
antibodies to neutralize these antigens (Leung, K. et
al., 2007).
The Artificial Immune Systems (AIS) are
adaptive systems which are inspired by human
immune system and offer similar features of
biological immune system such as self-organization,
noise tolerance and memory mechanism, while
having the ability to learn(J. H. Ang and al., 2010).
244
Daoudi R., Djemal K. and Benyettou A..
Digital Database for Screening Mammography Classification Using Improved Artificial Immune System Approaches.
DOI: 10.5220/0005079602440250
In Proceedings of the International Conference on Evolutionary Computation Theory and Applications (ECTA-2014), pages 244-250
ISBN: 978-989-758-052-9
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)
There are many models of Artificial Immune
Systems including negative selection, clonal
selection and immune networks.
The immune network theory was first proposed
by Jerne in 1947 (Jerne, N.K, 1974), the hypothesis
was that the immune system maintains an idiotypic
network of interconnected antibodies to recognize an
antigen (Aickelin et al., 2014). The negative
selection is a mechanism that protects the body from
self-reactive lymphocytes. It deals with the immune
system’s ability to detect unknown antigens while
not reacting self-cells (Daoudi et al., 2013). The
Clonal Selection algorithms are derives from the
clonal selection principle, which is based on
initiation of candidate solution, affinity maturation,
selection, cloning, mutation and reselection.
In the last decade, AIS have proven their
effectiveness in different areas and especially in the
medical field, they were applied to the detection of
lung disease, diagnosis of diabetes, tuberculosis,
heart disease and the detection of several types of
cancers... etc..
This paper proposes enhancements of a popular
clonal selection algorithm named CLONALG, for
classification of breast cancer cells into
benign/malignant classes taking into account three
new descriptors of the Digital Database for
Screening Mammography (DDSM). The results
obtained are compared to different AIS algorithms
and SVM.
2 DDSM CLASSIFICATION
METHOD
In This Section, we first present CLONALG
algorithm and it limitations, and then we give
detailed descriptions of the enhancements made on
this algorithm for breast cancer classification.
2.1 CLONALG Limitations
From proposed works respectively in (Daoudi et al.,
2013a) and (Daoudi et al., 2013b), we try to enhance
CLONALG algorithm. The first approach named
Cells Clonal Selection (CCS-AIS), the principle is to
select the best cells to be cloned by calculating the
averages of groups of the most competent cells. The
second one Medial Filter Artificial Immune System
(MF-AIS), the algorithm introduces median filter
principle to create the cell to be cloned.
Both algorithms propose improvements of
CLONALG algorithm (De Castro et al., 2002) which
is one of the most popular algorithms in the field of
the Artificial Immune systems using the principle of
the clonal selection.
We can distinguish two main limitations in
CLONALG algorithm. Indeed, the first limitation
we can observe is in the initialization step,
CLONALG algorithm takes a random population of
antibodies before launching the learning step. These
cells are selected randomly from the set of training
examples, which means that the initial cells do not
represent necessarily all of the cells to learn.
Learning will then depend on this set of randomly
initialized cells.
The second limitation of CLONALG is in the
training step, CLONALG select for each example to
learn a set of memory cells, clone and mutate these
cells; a reselection of the best mutated clones is
made thereafter to be added to the memory cells set.
Next, CLONALG replace P worst cells by randomly
created ones, even if the randomly generated cells
are worse than the rejected ones or if the rejected
cells can be more representatives of other training
examples in next generations, no check is made.
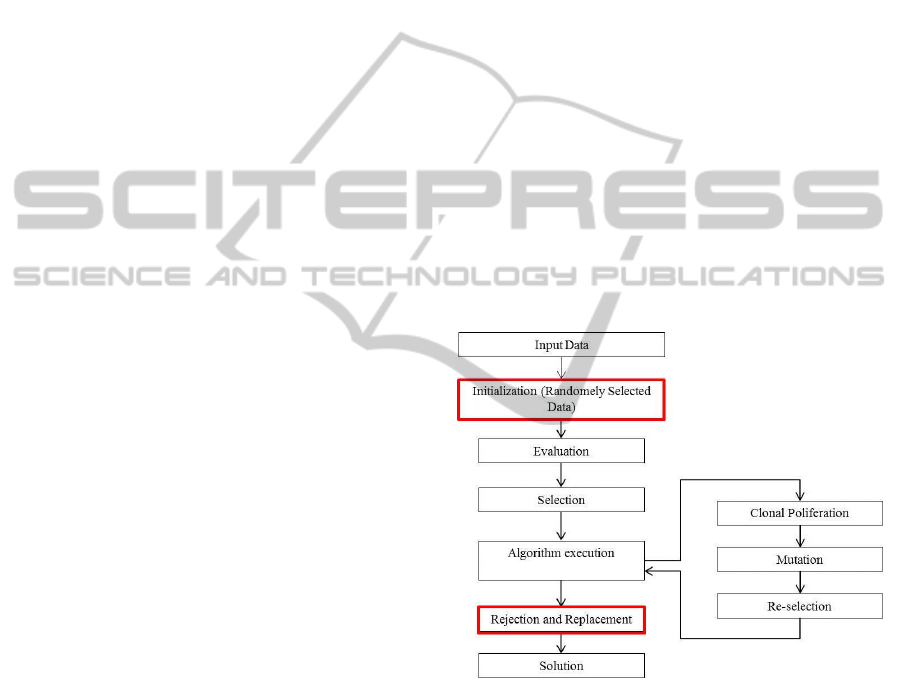
Figure 1 provides a simple flowchart of the
CLONALG algorithm.
Figure 1: Flowchart of CLONALG.
2.2 Cloned Cell Creation in
CLONALG System by CCS-AIS
and MF-AIS
The first improvement proposed in both proposed
techniques treats the problem of initialization of
antibodies before launching the learning, instead of
picking randomly some examples of training
database, the initialization step consists in dividing
every class of learning into subgroups, the average
cell of every local subgroup is considered as initial
antibody created to launch the learning algorithm.
DigitalDatabaseforScreeningMammographyClassificationUsingImprovedArtificialImmuneSystemApproaches
245
This process will allow the initial antibodies to
represent all the examples to be learnt and not some
only. Other various improvements brought to
CLONALG in both approaches are during the
learning phase, in the choice of the cell to be cloned.
In CCS-AIS, for each example to learn, the
selection of the closest cells is made and an average
cell of these cells is created, if the average cell has
better affinity than the nearest memory cell, it will
be added to the set of memory cells, and a set of the
best mutated clones of this average cell maximizing
the affinity with the learning example is selected to
join the set of memory cells, otherwise the best
antibody will undergo to the operators of cloning
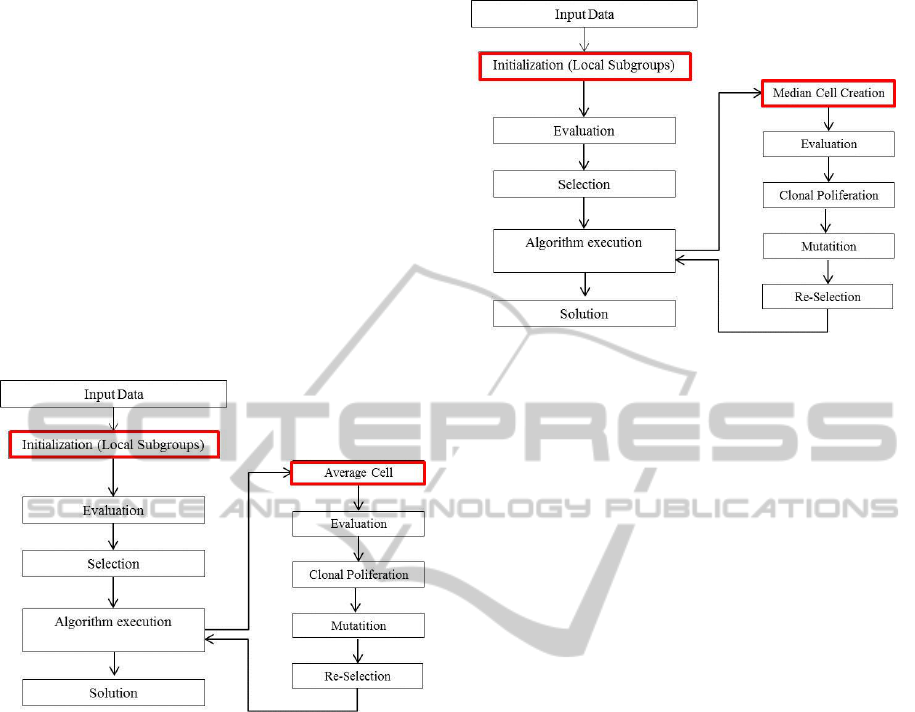
and mutation. Figure 2 presents the flowchart of the
CCS-AIS algorithm.
Figure 2: Flowchart of CCS-AIS.
In MF-AIS, the learning phase consists in
creating a cell that will be cloned and mutated
named median cell. Knowing that the learning base
is composed of N attributes, the process of creating
the median cell is done by selecting N closest cells
to the learning example by measuring affinity, and
taking the median value of each attribute from these
cells. The median cell created is subsequently
evaluated and if it has a higher affinity than the
nearest antibody, it will be added to all memory cells
as well as all its best mutated clones. The diagram of
MF-AIS is given in Figure3.
Figure 3: Flowchart of MF-AIS.
The classification step consists of comparing
each example to classify to all memory cells
obtained at the end of learning, and the example is
assigned to the class of the memory cell that
maximizes the affinity.
Both detailed above approaches have been
applied for the diagnosis of breast cancer on the
Wisconsin Breast Cancer Database (WBCD), and
the results were promising, to validate the
approaches, we propose in this article apply them on
another database widely used in the field of
detection of breast cancer using new descriptors, and
to compare the results obtained with other AIS
methods.
2.3 Improvements on DDSM
In order to compare the results obtained in our
previous work and validate our approaches, we
chose a database constructed from digitized films
named Digital Database for Screening
Mammography (DDSM). It was assembled by a
group of researchers from the University South
Florida and was completed in 1991 (M. Heath et al.,
2000). DDSM contains 2620 cases collected from
Massachusetts General Hospital, Wake Forest
University School of Medicine, Sacred Heart
Hospital and Washington University of St. Louis
School of Medicine.
Digital Database for Screening Mammography
was widely used by the scientific community in the
field of breast cancer; it has the advantage of using
the same lexicon standardized by the American
College of Radiology in BI-RADS. The different
patient records were made in the context of
screening and were classified into three cases:
normal case (no lesions), benign case and malignant
ECTA2014-InternationalConferenceonEvolutionaryComputationTheoryandApplications
246
case, each file is composed of four views containing
the mediolateral-oblique incidence (MLO) and the
Cranio-Caudal incidence (CC) of each breast. These
files are also provided with annotations provided by
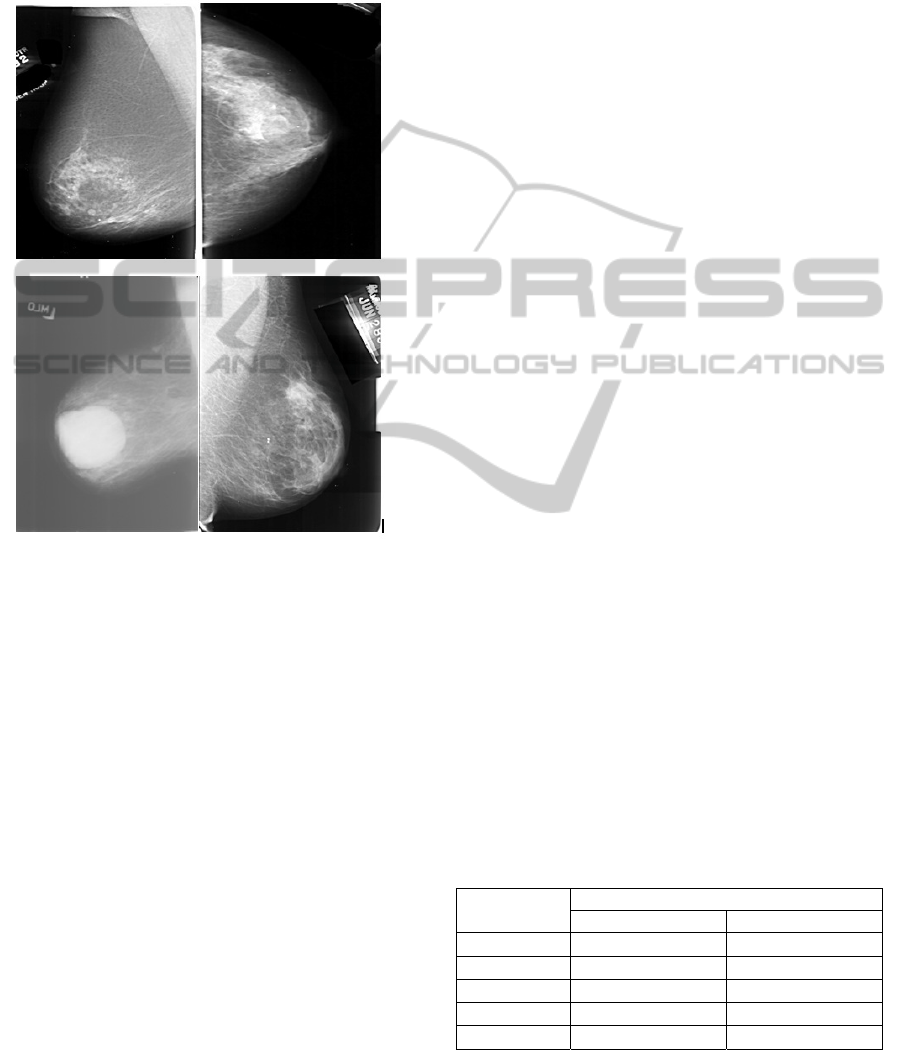
expert radiologists. Fig 4 shows samples DDSM
used in the evaluation.
Figure 4: Samples of DDSM database used in evaluation.
Sub-base of DDSM was created consisting of
242 masses: 128 benign and 114 malignant. These
examples will be partitioned into training examples
and test examples.
Part description of breast masses is a very
important part , in (Cheikhrouhou, 2012), the author
proposed three new descriptors: the skeleton
endpoint (SEP), Protuberance selection (PS) and the
spiculated mass Descriptor (SMD), which were
compared to 19 other descriptors proposed in the
literature including:
Area;
Perimeter;
Circularity;
Squareness;
Modified squareness;
Compactness;
Curvature;
Elliptic normalized skeleton;
Number of large protuberances and
depressions;
Average of the normalized radial length;
Standard deviation of the normalized radial
length;
Entropy;
Ratio of surface;
Roughness;
Rate of zero crossing;
Difference of the standard deviations;
Modified entropy;
Report of surface modified;
Rate of modified crossing in zero.
In this work, all of the 22 descriptors are used in
the stage of classification by both approaches
detailed in the section 2.1; the results obtained as
well as a comparison of the results are presented in
the following part.
3 RESULTS AND DISCUSSION
3.1 DDSM Classification Results
The performances of the approaches are studied
using DDSM, the training data are antigens
represented by feature vectors, also, the antibodies
have the same shape as the antigenic vectors, and the
Euclidean distance is used as a measure of
similarity; the 5-CV (Cross validation) is used.
Specifically, each dataset is split into five equal
subsets. In every run of each algorithm, one of the
subsets was used as test set, and the remaining four
comprised the training set. The average of 5 times of
successive runs is taken as classification result. After
1, 2, 5, 8 and 10 iterations, memory cells generated
at the end of learning are used in classification, by
comparing each cell to classify to all of the created
memory cells, and assigning it to the class
containing the memory cell with the highest measure
of similarity, simulation and implementation are
done using MATLAB 7.11.0. Tables 1 and 2
summarize the results obtained:
Table 1: Classification accuracies of Cells Clonal
Selection AIS on DDSM.
Iteration N° Classification Accuracies (%)
Train Test
1
72,85 71,21
2
93,24 96,48
5
93,56 96,81
8
93,74 96,69
10
97,70 98,13
DigitalDatabaseforScreeningMammographyClassificationUsingImprovedArtificialImmuneSystemApproaches
247
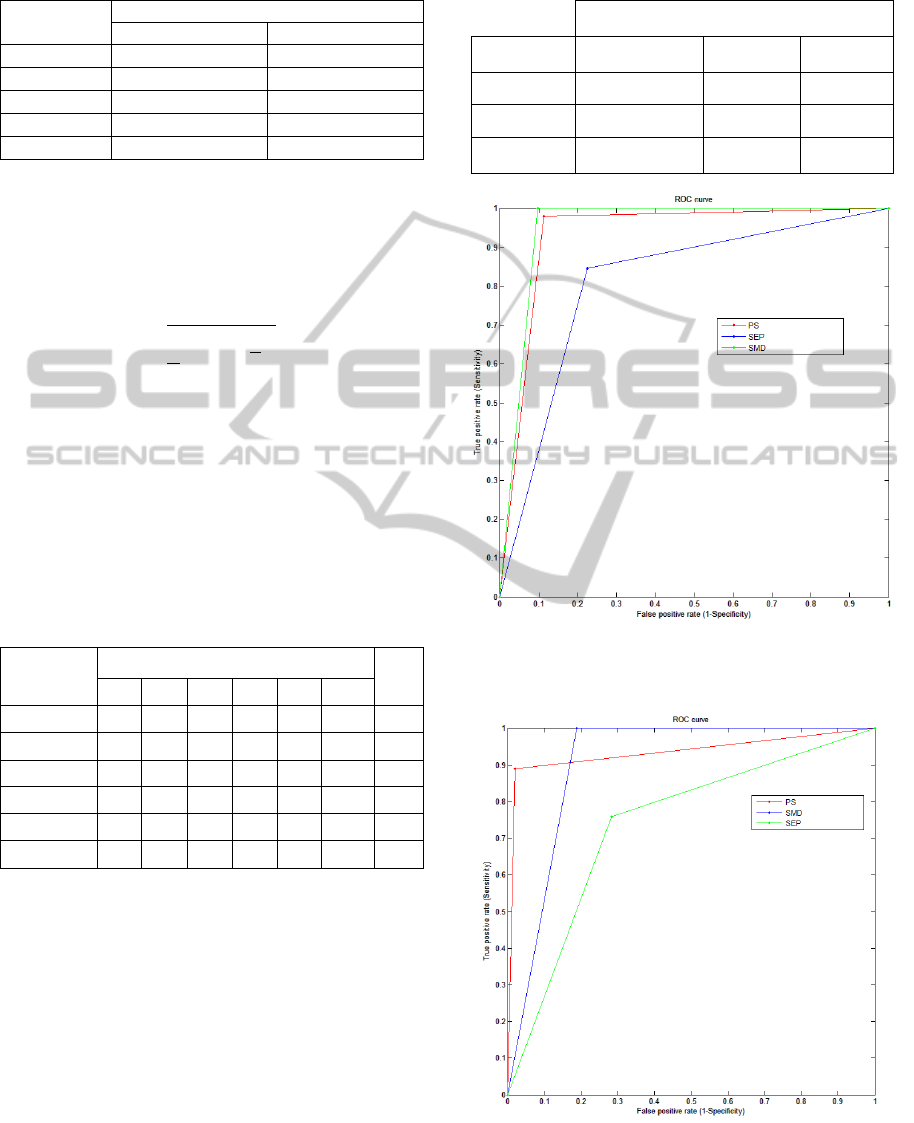
Table 2: Classification accuracies of Median Filter AIS on
DDSM.
Iteration N° Classification Accuracies (%)
Train Test
1
95,89 92,89
2
96.76 94.23
5
95,18 93,87
8
95,31 96,96
10
96,33 97.74
So as to compare the results obtained, we have
implemented different AIS algorithms and applied
them on DDSM using the same parameters; Table 3
shows the test results of five successive runs, the
average accuracy and supported by the standard
deviation value of each AIS classification algorithm:
1
²
1
(1)
Calculating the standard deviation (1) of
successive runs of each algorithm allows us to know
how the results of different executions are far from
the average accuracy. Indeed, for CCS-AIS de
standard deviation is 0.82 while it is 1.85 for
CLONALG algorithm.
Table 3: Classification accuracies of different AIS
algorithms on DDSM.
AIS
Algorithm
Classification Accuracies (%)
σ
(Standard
Deviation)
1
st
run 2
n
d
run 3
r
d
run 4
t
h
run 5
t
h
run Average
CSA 90.15 92.21 88.34 93.67 89.33 91.17 2.21
CLONALG 96.35 92.67 97.26 94.83 96.75 95.57 1.85
CLONAX 95.77 93.84 91.96 94.51 95.18 94.25 1.47
AIRS 81.27 83.54 80.12 84.67 81.16 82.15 1.88
CCS-AIS 98.33 99.09 97.92 96.87 98.46 98.13 0.82
MF-AIS 98.94 96.87 97.75 96.66 98.49 97.74 0.99
To validate the new proposed descriptors, author
in (Cheikhrouhou, 2012) classified each one
separately using SVM classifier, on order to
compare our approaches, we applied CCS-AIS AND
MF-AIS on the three new descriptors, the results
obtained are given in Table 4, and ROC curves
representing the results of the two proposed
approaches are presented un Figure 5 and Figure 6 .
Table 4: Classification accuracies of different AIS
algorithms on DDSM.
Classification Accuracies
Descriptors SVM
(Cheikhrouhou,2012)
CCS-AIS MF-AIS
SEP 92% 95.43 93.12
PS 93% 97.69 96.52
SMD 97% 98.95 98.57
Figure 5: Roc Curves of test results Cells Clonal
Selection-AIS on the three new descriptors proposed.
Figure 6: Roc Curves of test results of Median Filter-AIS
on the three new descriptors proposed.
ECTA2014-InternationalConferenceonEvolutionaryComputationTheoryandApplications
248

3.2 Discussion
From the tables above, we can see that the
improvements brought to CLONALG show
effective. The choice of the initial antibodies to
launch an AIS algorithm directly affects the results,
it is necessary that these antibodies represent all
learning classes and not just some examples only, it
will allow to find the cell which represents most
exactly the example to learn. The creation of local
subgroups from learning classes has treated this
problem in each of the both proposed approaches.
The creation of the cloning cell also played an
important role in the learning phase, in Cells Clonal
Selection AIS, the creation of this cell was done by
calculating an average cell of the best memory cells,
while in median Filter AIS, the median cell was
created from the median values of each attribute of
the matrix of the nearest memory cells, and to
maximize the affinity of each cell created, they are
compared to the best antibodies and they are added
to the set of memory cells only if they are better. No
cell can be representative in next generation was
rejected.
Classification results after 20 iterations on
DDSM taking into account three new descriptors
proposed in (Cheikhrouhou, 2012) are 98.71% for
cells clonal selection AIS and 98.21% for Median
Filter for AIS, efficient results compared to other
approaches AIS.
The classification results of each one of the new
descriptors separately prove that the proposed
approaches prove the effectiveness of the proposed
techniques comparing to the SVM classifier.
4 CONCLUSION
In this work, the classification of DDSM by Clonal
Selection-Based AIS was presented. Each of the two
techniques deals with the problem of initialization of
the antibodies before the launching of the training
Phase for the representation of the entirety of the
data to learn and proposes a new method for the
choice of the cell to clone in order to maximize
affinity with the training example. Cells Clonal
Selection AIS proposes the creation of an average
cell from the closest memory cells, and Median
Filter AIS introduces the principle of the median
filter to create a median cell for the cloning. The
obtained results indicate that the improvements
brought to CLONALG are effective and can be of a
precious help to the experts for a second opinion in
their diagnosis of breast cancer.
Note that we have made is that the two
approaches in particular and AIS algorithms
generally require heavy computation time , our next
work will focus on the treatment of this problem.
REFERENCES
Ferlay J, and al., 2013. GLOBOCAN 2012 v1.0, Cancer
Incidence and Mortality Worldwide: IARC
CancerBase No. 11 [Internet]. Lyon, France:
International Agency for Research on
Cancer.Available from http://globocan.iarc.fr
Marcano-Cedeno, A., J. Quintanilla-Dominguez, and D.
Andina,2011,WBCD breast cancer database
classification applying artificial metaplasticity neural
network. Expert Systems with Applications, 38(8): pp.
9573-9579.
Timmy Manning and Paul Walsh, 2013.Improving the
Performance of CGPANN for Breast Cancer
Diagnosis Using Crossover and Radial Basis
Functions, in Proceeding of 11th European
Conference, EvoBIO 2013, Vienna, Austria.
Hossein Ghayoumi Zadehand al.,2012.Diagnosis of Breast
Cancer using a Combination of Genetic Algorithm and
Artificial Neural Network in Medical Infrared Thermal
Imaging, Iranian Journal of Medical Physics Vol. 9,
No. 4, pp 265-274.
Aboul Ella Hassanien and al. 2014. MRI breast cancer
diagnosis hybrid approach using adaptive ant-based
segmentation and multilayer perceptron neural
networks classifier, Elsevier Applied Soft Computing ,
Volume 14 , pp 62–71
Mahnaz Rafie and Ali Broumandnia, 2013.Evaluation of
Cancer Classification Using Combined Algorithms
with Support Vector Machines, International Journal
of Computer & Information Technologies (IJOCIT13),
Vol 1, Issue 2 , pp 137-148
Aboul Ella Hassanien and al.2013.Breast Cancer
Detection and Classification Using Support Vector
Machines and Pulse Coupled Neural Network, in
Proceedings of the Third International Conference on
Intelligent Human Computer Interaction (IHCI 2011),
Prague, Czech Republic, Advances in Intelligent
Systems and Computing Volume 179, 2013, pp 269-
279
Jain, R. and J. Mazumdar, 2003. A genetic algorithm
based nearest neighbor classification to breast cancer
diagnosis. Australasian physical & engineering
sciences in medicine / supported by the Australasian
College of Physical Scientists in Medicine and the
Australasian Association of Physical Sciences in
Medicine, 26(1): p. 6-11.
Mazurowski, M.A., et al.2007. Case-base reduction for a
computer assisted breast cancer detection system using
genetic algorithms. 2007 Ieee Congress on
Evolutionary Computation, Vols 1-10,
Proceedings2007. 600-605.
DigitalDatabaseforScreeningMammographyClassificationUsingImprovedArtificialImmuneSystemApproaches
249

Wan Noor Aziezan Baharuddin and al. 2013. Mamdani-
Fuzzy Expert System for BIRADS Breast Cancer
Determination Based on Mammogram Images,
Springer Soft Computing Applications and Intelligent
Systems Communications in Computer and
Information Science Volume 378, pp 99-110
Anuarg Sharma and Dharmendra Sharma, 2011. Clonal
Selection Algorithm for Classification , in Proceeding
of 10th International Conference on Artificial Immune
Systems ICARIS 2011, Vol 6825 of LNCS,(pp. 361-
370). Springer.
Rima Daoudi, Khalifa Djemal and Abdelkader Benyettou,
2013a. Cells clonal selection for Breast Cancer
classification, in proceeding of 10th International
Multi-Conference on Systems, Signals & Devices
SSD13.
Rima Daoudi, Khalifa Djemal and Abdelkader Benyettou.
2013b. An Immune-Inspired Approach for Breast
Cancer Classification, in proceeding of 14th
International Conference on Engineering
Applications of Neural Networks EANN 2013, Series
Volume 38, pp 273-281, Springer.
Leung, K., Cheong, F., Cheong, C.2007. Generating
compact classifier systems using a simple artificial
immune system. IEEE Transactions on Systems Man
and Cybernetics Part B-Cybernetics 37(5), 1344–1356
(2007)
J. H. Ang, K. C. Tan, A. A. Mamum, 2010. An
evolutionary memetic algorithm for rule extraction,
Expert Systems with Applications 37 (2010) 1302–
1315
Jerne, N.K., 1974, Towards a Network Theory Of
Immune System. Annales D Immunologie,.C125(1-2):
p. 373-389.
Uwe Aickelin, Dipankar Dasgupta, Feng Gu,2014.
Artificial Immune Systems, Search Methodologies,
Springer Science+Business Media New York, pp 187-
211,
De Castro, L. N.; Von Zuben, F. J,2002.Learning and
Optimization Using the Clonal Selection Principle.
IEEE Transactions on Evolutionary Computation,
Special Issue on Artificial Immune Systems (IEEE) ,
2002. 6. (3): 239–251.
M. Heath, K. W. Bowyer, D. Kopans, et al. 2002. The
Digital Database for Screening Mammography,
presented at 5th International Workshop on Digital
Mammography Toronto, Canada, 2000.
Imene Cheikhrouhou Kachouri, 2012, Description et
classification des masses mammaires pour le
diagnostic du cancer du sein, Ph.D. Thesis. Uiversity
of Evry Val d’Essone: France.
ECTA2014-InternationalConferenceonEvolutionaryComputationTheoryandApplications
250
