
Disease Identification in Electronic Health Records
An Ontology based Approach
Ioana Barbantan, Camelia Lemnaru and Rodica Potolea
Computer Science Department, Technical University of Cluj-Napoca, Gh. Baritiu St, 28, Cluj-Napoca, Romania
Keywords: Electronic Health Records, Concept Annotation, Ontology, Negation, Prefixes, Structure.
Abstract: Exploiting efficiently medical data from Electronic Health Records (EHRs) is a current joint research focus
of the knowledge extraction and the medical communities. EHR structuring is essential for the efficient
exploitation of the information they capture. To that end, concept identification and categorization represent
key tasks. This paper presents a disease identification approach which applies several NLP document pre-
processing steps, queries the SNOMED-CT ontology and then applies a filtering rule on the retrieved
information. The hierarchical approach provides a better filtering of the concepts, reducing the amount of
falsely identified disease concepts. We have performed a series of evaluations on the Medline abstracts
dataset. The results obtained so far are promising – our method achieves a precision of 87.79% and a recall
of 87.12%, better than the results obtained by Apache’s cTAKES system on the same task and dataset.
1 INTRODUCTION
The 21
st
century technological revolution has had a
great impact on our everyday life, by making it
easier for us to communicate, organize, access
information, and so on; it has also transformed the
way we handle our health – we are quite accustomed
to searching online for any symptoms we might be
experiencing and establishing a diagnosis, and
(perhaps) even a treatment schema, on our own. But
online information is not always reliable.
Wearable technology is likely to transform
medical care, by helping both patients and clinicians
monitor vital signs and symptoms. Systems which
track the activity of elderly people and send health
measurements to their caregivers or those which
measure and send various body values to the
patients’ doctors are already established on the
market.
Consequently, Electronic Health Records
(EHRs) are adopted by an accelerated increasing
number of medical doctors, pharmaceutical
companies, caregivers and personal trainers. EHRs
represent a step forward in the development of the
medical system, by capturing the medical history
and current patient conditions with detailed
information about symptoms, procedures,
medications, illnesses or allergies. They are an
important source of knowledge if exploited
correctly: one can extract information on disease
interactions, the influence of demographics on
patient conditions, and so on. But, in order to do
this, the documents need to be clear, unambiguous
and should carry correct information. In most cases,
EHRs are unstructured and may contain recurrent
information.
Therefore, the final goal of our work is to
perform a structuring of the EHRs and further design
personal medical assistant applications (fig. 1). The
benefits of such applications are manifold: a shorter,
less painful and less expensive diagnosis process;
assist patients when they require additional
information regarding their condition; monitor and
transmit (and alert) health state; provide easier
access to medical information for physicians.
The flow in fig. 1 depicts the two main steps to
consider in order to reach the desired outcomes:
EHR structuring and knowledge extraction. We are
currently focusing on EHR structuring, and in this
paper we tackle an essential task for this step:
automatic concept annotation, with a focus on
disease annotation, and propose an ontology based
disease identification approach.
The rest of the paper is organized as follows: the
next section discusses related approaches. Section 3
sets the background of our research. In section 4 we
present our vision on concept identification in EHRs
261
Barbantan I., Lemnaru C. and Potolea R..
Disease Identification in Electronic Health Records - An Ontology based Approach.
DOI: 10.5220/0005082002610268
In Proceedings of the International Conference on Knowledge Discovery and Information Retrieval (KDIR-2014), pages 261-268
ISBN: 978-989-758-048-2
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

Figure 1: Flow for knowledge extraction from medical documents.
with focus on the ontology based approach proposed
in this paper. Section 5 presents the experiments
performed and a discussion based on the results
obtained. The last section presents the concluding
remarks.
2 RELATED WORK
In (Sibanda, 2006), the authors describe a statistical
semantic category recognizer for discharge
summaries, which employs a multiclass SVM
classifier on a set of orthographic, lexical, syntactic
and ontological features.
The authors focus on 8 semantic categories and
show that, for clinical text, the lexical and syntactic
contextual clues provide stronger indications of the
semantic category of a term than information
extracted from the UMLS (Unified Medical
Language System) Meta-thesaurus.
The authors of (Rosario, 2004) explore several
generative graphical models (both static and
dynamic) and an artificial neural network for the
task of semantic relation classification in bioscience
texts. Seven different relation types between entities
of the type treatment and disease are considered, and
a set of lexical, syntactic and semantic features. The
results reported by the authors show that the neural
network achieves superior recognition rates to the
graphical models. In the same area, (Rink, 2011)
proposes an approach for extracting relations
between medical problems, treatments, and tests in
clinical texts, by using a linear SVM classifier and a
rich set of features related to context, similarity,
nested relations, single concept, Wikipedia and
vicinity. The technique achieved the highest F1
score on the relation identification task in the 2010
i2b2 Challenge.
The NLP-SNOMED (Hina, 2010) system is a
rule based system which employs GATE (General
Architecture for Text Engineering) and SNOMED-
CT to annotate the key medical concepts in
discharge summaries. A strategy which aims to
extract and code diseases and procedures from
discharge summaries using the structure of the
summary to locate the appropriate text, divide text
segments which might contain disease data into
phrases, perform normalization and coding on the
phrases by using UMLS to find the concepts is
presented in (Long, 2005). In the evaluations
performed, the approach has managed to code all but
10 phrases out of 250 phrases to be coded, with 19
false positives. The system was further developed to
produce a list of concepts to be used by physician
annotators to speed the process of generating disease
and procedure lists for ICU cases in (Long, 2007),
with a reported recall of 93%, but a rather high
number of false positives.
In (Batool, 2013), the authors propose a system
which extracts medical terms from discharge
summaries and converts them into SNOMED-CT
codes, by combining several NLP pre-processing
techniques and an additional ontology and a
synonymy service to enhance recognition and
mapping to SNOMED-CT concepts. The authors
perform some evaluations on their approach, but do
not report absolute performance values since that
part is currently ongoing.
A regular expression parser which employs a
set of manually defined parsing rules to extract
medication information from discharge summaries is
presented in (Gold, 2008), with a reported precision
of 94% and recall of 83%.
Medical concept identification along with
negation and document structuring are presented in
cTAKES Apache clinical Text Analysis and
Knowledge Extraction System (Savona, 2010). It
relies on UMLS and medical ontologies – such as
SNOMED to identify diseases, symptoms or
procedures, and RxNorm (Nelson, 2011) to identify
drug names and specific components. The cTAKES
system consists of several modules: sentence
boundary detector, tokenizer, part-of-speech tagger,
negation identification, concept mapping, shallow
parser and named entity recognizer. The authors
have evaluated separately each module, reporting
F1-score values ranging from 0.58 to 0.957. The
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
262

named entity recognizer achieved an F-score of
0.715 for exact and 0.824 for overlapping spans.
3 BACKGROUND
This section attempts to set the background of the
work presented in this paper by introducing the main
concepts we operate with: EHRs and the need to
structure them, medical concept annotation as a step
in EHR structuring and the role of handling negation
in EHR concept identification and structuring.
3.1 Electronic Health Records
The EHRs are legal documents and must conform to
privacy and confidentiality policies. They capture
the patient’s consent and authorization for medical
procedures and information sharing with third
parties. A clinical discharge document in raw format
informs about document structuring into chapters
containing grouped information regarding:
Symptoms, Diseases, Diagnosis, patient’s Historical
information, Medical procedures (Long, 2005),
Medication (Halgrim, 2011), Investigations,
Demographic data or Follow-up information (Rudd,
2010).
EHRs focus on all medical aspects of a patient’s
health and help find correlations between the current
condition and previous investigations and conditions
(Clay, 2012). In most cases, EHRs are unstructured,
which means – among other things, that information
regarding a certain aspect may be found in several
document sections. In order to access information
efficiently and fast, it becomes imperative that all
documents are aligned to a standard structure.
3.2 Extracting Concepts from Medical
Documents
The end goal of our work is to obtain a semi-
supervised approach for assisted diagnosis,
procedures, treatment, based on the symptoms and
investigations performed. Thus, the first step in
structuring the EHRs is to identify the concepts and
the relations between them. Then, the following
combinations will be considered (see also fig. 2):
(symptoms - diagnosis), (symptoms - procedures),
(diagnosis - evolution) and (diagnosis - treatment).
Starting from the list of symptoms that a patient
experiences and those which he/she denies, the
system will be able to recommend diagnosis or
suggest several procedures to be carried out. Once
the diagnosis is established by the physician, the
system could recommend a treatment plan or
determine the disease progression.
Establishing these relationships requires that the
medical concepts are clearly and correctly identified
and annotated in the documents. When the
annotation is performed, the sentences containing
medical concepts are assigned to categories such that
all sentences regarding symptoms are found in the
symptoms section, the procedures related statements
are in the procedures section and so on, facilitating
the access to the information.
Two main approaches exist for extracting
entities from documents: using a set of regular
expressions to perform direct matching, or using a
machine learning classification methodology. Both
approaches require the existence of some auxiliary
resources such as dictionaries or ontologies which
are queried at some point.
Systematized Nomenclature of Medicine --
Clinical Terms (SNOMED-CT) represents a
clinical healthcare terminology (aligned to
international standards) designed for being used in
EHRs. It can be used to describe a patient’s
condition, the procedures performed, the spread of
epidemics and many more. It consists of more than
311,000 active concepts with unique meanings
organized into hierarchies from the most general to
the most specific concept. Each concept is assigned
a unique ID. In order to handle the synonymy of
concepts, SNOMED-CT uses descriptions for each
synonym of the concept. SNOMED-CT is
represented in a hierarchical form containing
grouped information about disorders, procedures or
body structures for identifying anatomical structures
affected, staging and scales to identify for example a
tumour staging.
A concept in SNOMED is represented by its
name and (possibly) alternative names, definition,
parent relationship and several IDs that help in the
unique identification of the concept in different
storage places. The information captured in
SNOMED is represented in RDF format, using basic
graph pattern triples <subject-predicate-relation>
(SPARQL, 2013) In order to query the ontology, the
SPARQL query language is employed. The queries
performed using SPARQL allow searching for
concepts by names, unique IDs or properties, for
discovering the relationships between concepts, and
also for result filtering. The concepts are related
based on the is-a relationship and a concept can have
several parents. Like in the case of Acute
appendicitis with peritoneal abscess is-a Acute
digestive system disorder which is-a Acute disease.
DiseaseIdentificationinElectronicHealthRecords-AnOntologybasedApproach
263
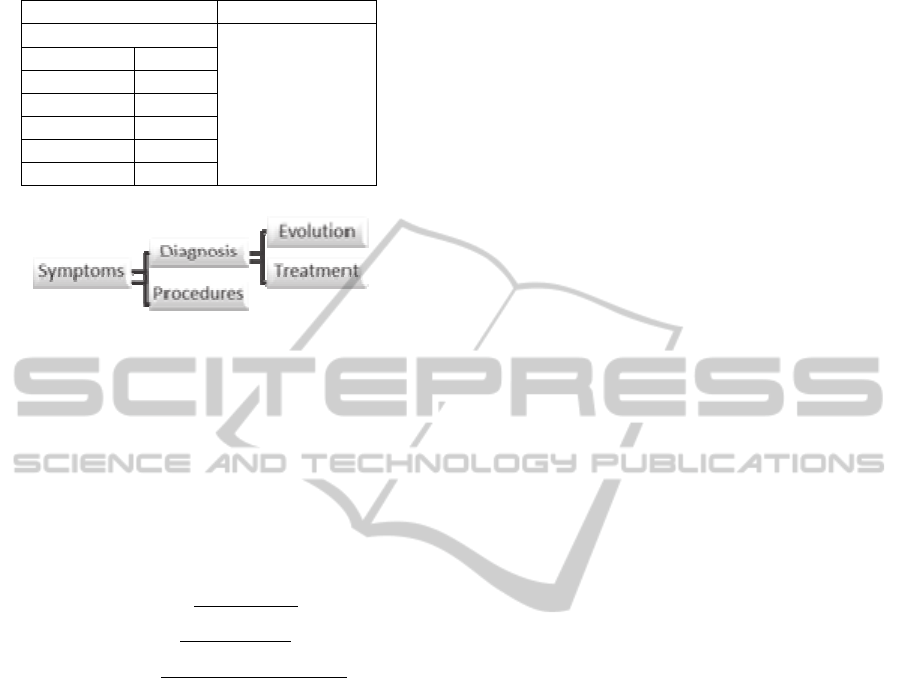
Table 1: Negation statistics for MTsamples dataset.
Medical concepts Common words
92.04%
7.96%
Sympto
m
45%
Dia
g
nosis 17.78%
Procedure 12.98%
Histor
y
1.92%
Medication 4.34%
Othe
r
15.59%
Figure 2: Relations between medical concepts.
3.3 Negation
As indicated by analyses performed on medical
documents, negative polarity sentences are rather
frequent in medical records: 2% of the concepts
have their value flipped due to negation (Barbantan,
2014b).
For example the following three sentences state
the same thing:
The patient has no symptoms.
The patient is asymptomatic.
The patient doesn’t have symptoms.
Thus, negation can be expressed using explicit
terms like no and n’t, but can also be expressed
using prefixes, such as a. In (Givon, 1993), explicit
negation is referred to as syntactic negation, whereas
negation with prefixes is termed as morphologic.
Studies on negation (Mutalik, 2001) (Councill,
2010) focus on syntactic negation alone.
However, analyses performed on the
MTSamples medical documents (MTSamples, 2012)
have revealed that morphologic negation is as
important (56% of the total number of negations is
morphologic, and 44% is syntactic).
Also, Table 1 presents a statistic performed on
the categories of concepts which appear negated in
the same dataset: 92% of the negations are related to
the medical concepts while only 7.96% are related to
common words. The most common negated medical
concepts are the symptoms – 45% of all negated
medical concepts. Therefore, in order to extract the
correct information from medical documents, it is
essential to separate between affirmed and negated
concepts: i.e. for establishing a diagnosis, the
affirmed symptoms are used to determine possible
diseases, whereas negated symptoms are employed
to refine that list via exclusion using the negated
symptoms.
However, negation analysis is no trivial task,
since the influence of negation identifiers can spread
to several parts of a sentence and change the
meaning of several concepts (as in “The patient did
not present with fever, headache or ocular pain”).
4 METHODOLOGY FOR
CONCEPT IDENTIFICATION
FROM EHRS
In our work so far, we have implemented several
algorithms for identifying medical concepts (needed
to further extract the categories). In (Barbantan,
2014a), we employed a vocabulary of terms and a
binary bag of words feature vector. In (Barbantan,
2014b), we also exploited the meaning of the terms.
Our current work proposes a more in depth
analysis of the concepts as we include the
relationships between concepts and their meanings,
by using well-established medical domain ontology.
4.1 Vocabulary Based Concept
Identification
In (Barbantan, 2014a), we have presented the BOW-
NPI methodology for negation identification using a
rule-based approach and a dictionary represented as
a bag of words. Negated concepts were identified by
consulting the NegEx list of negation identifiers
(Chapman, 2001). To deal with the morphologic
negation we employed a bag of words classification
approach where part of the corpus was used to create
the dictionary and the rest was used for testing. To
determine whether a word is negated with prefix, we
computed its validity by determining its existence in
the feature set. Using this approach we achieved a
precision of 95.79% and recall of 87.63%.
4.2 Dictionary Based Concept
Identification
The dictionary based approach (Barbantan, 2014b)
exploits the meaning of the words by using an
English language dictionary; negated compound
words were addressed by using an n-gram based
approach. The rules for negation identification used
in this approach are:
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
264
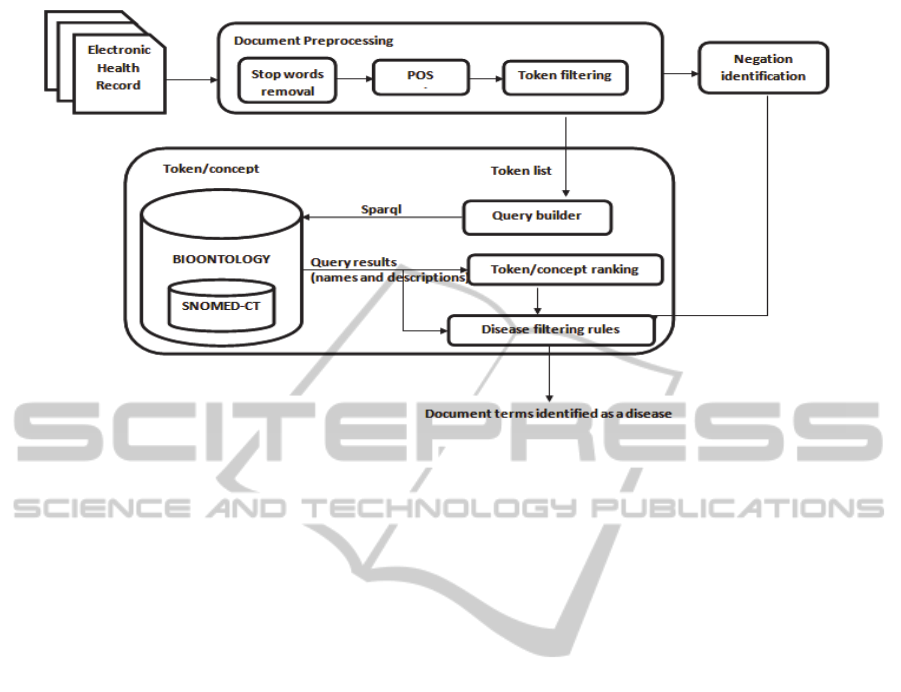
Figure 3: Disease identification flow.
Definition recurrence rule: the root of a prefixed
word is contained in the prefixed word’s definition.
Definition content rule: both the root of the
prefixed word and the prefix word are defined in
WordNet and the definition of the prefixed word
contains a negation identifier.
Hyphen rule: the prefix is followed by hyphen or
space – the case is handled by removing the special
character and sending the entity to be analysed with
the previous rules.
Compound words: progressively build a word from
consecutive letters on an n-gram basis; remove the
prefix and perform an analysis of the root. If the
word can be split into two words with definitions in
WordNet, we consider the word negated with
negation prefix. The precision achieved by this
approach was 95.96% and the recall 94.23%, on a
subset of EHRs provided by (MTsamples, 2012),
which yields an absolute improvement of 6.6% in
terms of recall and a small increment (0.17%) in
precision.
4.3 Ontology Based Concept
Identification
In this paper we attempt to improve our medical
concept identification approach by making use of a
specialized medical domain ontology. Using an
ontology to extract concepts from text provides
several advantages over any dictionary-based
approach. This is due to the fact that, like the
majority of words in natural language, medical
concepts can be expressed using several
terminologies. For example, in order to refer to a
respiratory manifestation, the medical doctors use
the concept “influenza”, in ICD-10 (WHO, 2004)
disease is identified by the J11.1 diagnosis code and
in common language we refer to it as “flu”.
Ontologies, unlike vocabularies or dictionaries, can
easily capture this aspect by means of relations, like
synonymy, hierarchical levels or different labels.
The proposed methodology is presented in fig. 3. It
works as follows: first, a series of pre-processing
steps are applied, where we remove the stop words
and parse the text into individual tokens. The tokens
are then submitted to the POS tagger and pronouns
are eliminated. For each remaining token, we decide
whether it is affirmed or negated via the negation
identification module; then, we query the ontology
and analyse the response. For this, we integrated the
web service provided by the SPARQL BioPortal
(Salvadores, 2012). In case a positive response is
presented as output, we determine whether this
response is related to a disease (to be described
shortly).
Initially, the query considered the name,
description and label of the concepts as they are
stored in the ontology. When evaluating the results
of the query, we noticed a relatively large number of
false positives. In order to remove such errors, we
introduced a supplementary condition which exploits
the hierarchical representation of the concepts in
SNOMED. As most of the diseases we search for in
the documents are actually leaf instances in the
ontology, we establish that a concept is a disease if
both the instance and its parent are diseases.
DiseaseIdentificationinElectronicHealthRecords-AnOntologybasedApproach
265

The SPARQL query we employ currently is:
PREFIX rdfs:
<http://www.w3.org/2000/01/rdf-schema#>
SELECT DISTINCT *
FROM
<http://bioportal.bioontology.org/ontol
ogies/SNOMEDCT>
WHERE {
?x rdfs:label ?label.
?x rdfs:subClassOf ?parent.
?parent rdfs:label ?parentLabel.
FILTER ((CONTAINS ( str(?label),
concept ) && (CONTAINS (
str(?parentLabel), concept)
};
Results post-processing: After determining the
possible disease concepts (via the rule stated above),
we apply a post-processing step which considers the
position of the concept in the retrieved response, in
order to finally establish whether the concept is
actually a disease. We split the description of the
disease into sections and define as leading concepts
the tokens found in the first positions. The concept is
considered to be a disease when its occurrences as
leading concept outnumber the occurrences in the
final terms of the description. Otherwise, the word is
considered as auxiliary term in the description of
some disease. The pseudo-code of the post-
processing procedure is presented below:
Procedure FilterResults
Input: Token – the current token, the
subject of the current query
Results – a set of strings,
respresenting the output returned by
the current query
Output: IsDisease – a boolean, TRUE if
the current token represents a disease
concept
Procedure:
leading <- 0
for result in Results
resultArr <- result.split()
for i<-1 to resultArr.size()/2
if(resultArr[i]=Token) then
leading <- leading + 1
endif
endfor
if(leading≥Results.size()-leading)
then
IsDisease <- true
else
IsDisease <- false
endif
return IsDisease
To give an example of how the approach works,
say we want to determine whether the word
influenza is a disease, one of the responses obtained
is represented by the concept with SNOMED ID
81524006, associated with the following description
“Influenza due to Influenza virus, type C (disorder)”.
The token influenza is a leading concept as it is on
the first position in the disease description. But, we
obtain the same result when performing an
interrogation for the token virus. However, in this
case virus is not a leading concept and therefore it is
not annotated as a disease.
A medical concept can match a disease, a
procedure, a body structure or a situation. As our
goal is to identify diseases, we remove the cases
when the concept is related to hierarchies which
don’t contain diseases, and verify whether the
concept can match the name provided for the
disease.
5 EXPERIMENTAL WORK
This section presents the experiments performed so
far on the proposed disease identification method.
Since there are no publicly available annotated
EHRs (to the best of our knowledge), for now we
validate our approach on medical documents with
similar content, although possibly a different layout:
the annotated Medline abstracts dataset.
5.1 Medline Abstracts Dataset
The U.S. National Library of Medicine contains a
collection of biomedical abstracts and citations
which are constantly updated. Part of these abstracts
were previously annotated and employed in the
identification of relations between medical concepts
like diseases and treatment in (Rosario, 2004). The
annotation process was performed by a student with
biological background. The labelled data used in our
analysis consists of more than 120 abstracts. The
identified concepts were surrounded by tags related
to the concept’s type like <DIS>, <DISEASE> or
<DIS_VAG> for diseases and <TREAT>,
<TREATONLY> corresponding to treatment.
5.2 Experimental Setup
In order to evaluate the efficiency of our approach,
we used a subset of abstracts from the annotated
abstracts. We considered only the diseases that were
clearly annotated and ignored the cases where
vagueness was induced, such as <DIS_VAG> tags.
To prepare the dataset, we removed the tags related
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
266

to diseases. The diseases appear in the abstracts
either as nouns (mostly) or as ICD codes.
We have performed the same disease
identification task using Apache’s cTAKES
(Savona, 2010) module for named entity
recognition.
5.3 Results and Discussion
For the specific classification task we are focusing
on, both recall and precision are important to
establish the performance. Thus, we report both
values in Table 2. As it can be observed, we have
performed several analyses of the performance of
our approach: using the initial ontology strategy
query, which did not exploit hierarchy-related
information (Initial in table), then recomputed the
values taking into account only the diseases which
appeared in SNOMED (Initial,
SNOMED diseases
only), and using the hierarchy information as well
(Initial+Hierarchy).
The rather modest value obtained for recall in E1
is partly motivated by the fact that 29% of the
diseases which appeared in the documents had no
identifiers in SNOMED. If we consider only the
diseases that are represented in SNOMED (E2), we
obtain a recall value of 66.25%. Using also the rule
which exploits the parent relationship as defined in
the ontology, we obtained significantly better results
for both precision (89.79%) and recall (87.12%).
Table 2: Disease identification performance on the
Medline abstracts dataset.
Experiments Precision Recall
E1: Initial 84.12% 47.31%
E2: Initial, SNOMED
diseases only
84.12% 66.25%
E3: Initial+Hierarchy
89.79% 87.12%
Improvement E1 -> E3 5.67% 39.99%
cTAKES 63.51% 78.33%
The analysis of the missed disease concepts
during identification yielded a series of issues which
can be addressed at three different levels: dataset
level, word level and ontology level.
At the dataset level, the annotations are not
always consistent; for example, we find cases when
the entire concept “prostate cancer” is annotated and
cases when only “cancer” is annotated as a disease.
Also, at word level (but associated with the
dataset), misspellings are fairly common – for
example we found the disease Alzheimer spelled as
“Alzeimer”, which is why this concept was not
identified as a disease. To solve this issue we
propose using a spell checking algorithm on the
documents in the pre-processing step and
performing a non-exact matching of the concepts in
the documents with the entities in the ontology (use
a similarity measure instead of an exact match).
At ontology level, the issues we identified are
related to the degree of synonymy offered by the
ontology. Some of the concepts are identified only
by their medical representation – for example, the
Down syndrome is represented in the ontology as
“trisomy”.
6 CONCLUSIONS
This paper continues our research efforts in
structuring EHRs, by proposing an approach for
identifying diseases in unstructured discharge
summaries. The method employs the SNOMED-CT
medical ontology to identify diseases in the medical
documents. It consists of a series of text pre-
processing steps, followed by the actual
identification, in which the ontology is queried and
the results are processed by a set of rules to
determine whether the tokens (or the list of tokens)
in the query represent a disease or not. In the
evaluations performed we have compared the
performance of our approach with that achieved by a
similar system – cTAKES significantly better recall
and precision values, thus we can claim that our
approach is indeed promising.
Also, we have identified a set of issues at
different levels: dataset, word and ontology, and we
are in the process of investigating several tactics for
addressing them, such as including auxiliary
resources (e.g. a synonymy service) or performing a
similarity based matching between the concepts.
As further work, we propose exploiting all the
properties defined in the SNOMED ontology in
order to identify all types of medical concepts which
may appear in EHRs: diseases, procedures,
symptoms, body structures.
ACKNOWLEDGEMENT
The authors would like to acknowledge the
contribution of the COST Action IC1303 -
AAPELE.
The work presented in this paper was partially
supported by the Post‐Doctoral Programme
POSDRU/159/1.5/S/ 137516, project co‐funded
from European Social Fund through the Human
DiseaseIdentificationinElectronicHealthRecords-AnOntologybasedApproach
267

Resources Sectorial Operational Program
2007‐2013.
REFERENCES
Barbantan, I., Potolea, R. 2014a. Towards knowledge
extraction from electronic health records - automatic
negation identification. International Conference on
Advancements of Medicine and Health Care through
Techonology.”, Cluj-Napoca, Romania.
Barbantan, I., Potolea, R., 2014b. Exploiting Word
Meaning for Negation Identification in Electronic
Health Records, IEEE AQTR, Cluj-Napoca, Romania.
Batool, R., et al, 2013. Automatic extraction and mapping
of discharge summary's concepts into SNOMED CT.
Annual International Conference of the IEEE
Engineering in Medicine and Biology Society.
Chapman, W., Bridewell, W., Hanbury, P., Cooper, G.F.,
Buchanan, B.G., 2001. A Simple Algorithm for
Identifying Negated Findings and Diseases in
Discharge Summaries. Journal of Biomedical
Informatics 34(5): 301-310.
Clay, R. A., 2012. The Advantages of Electronic Health
Records. American Psychological Association,
STATE LEADERSHIP CONFERENCE. 43: 72.
Councill, I. G., McDonald, R., Velikovich, L., 2010.
What’s great and what’s not: learning to classify the
scope of negation for improved sentiment analysis.
Proceedings of the Workshop on Negation and
Speculation in Natural Language Processing, Uppsala.
Givon, T., 1993. English Grammar: A Function-Based
Introduction. Benjamins, Amsterdam, NL.
Gold, S., Elhadad, N., Zhu, X., Cimino, J.J., Hripcsak, G.,
2008. Extracting Structured Medication Event
Information from Discharge Summaries. D. o. B. I.
Department of Biomedical Informatics. New York.
Halgrim, S. R., Xia, F., Cadag, E., & Uzuner, Ö, 2011. A
cascade of classifiers for extracting medication
information from discharge summaries. Journal of
Biomedical Semantics.
Hina, S., 2010. Extracting the Concepts in Clinical
Documents Using SNOMED-CT and GATE. Fourth
i2b2/VA Shared-Task and Workshop Challenges in
Natural Language Processing for Clinical Data.
Long, W., 2005. Extracting Diagnoses from Discharge
Summaries. AMIA 2005 Symposium Proceedings:
470-474.
Long, W., 2007. Lessons extracting diseases from
discharge summaries. AMIA Annual Symposium
Proceedings.
MTsamples. "Transcribed Medical Transcription Sample
Reports and Examples." Last accessed on 23.10, 2012.
Mutalik, P.G., Nadkarni, P.M. 2001. Use of general-
purpose negation detection to augment concept
indexing of medical documents: a quantitative study
using the UMLS. Journal of the American Medical
Informatics Association 8: 598-609.
Nelson, S.J., Zeng, K., Kilbourne, J., Powell, T. and
Moore, R. 2011. Normalized names for clinical drugs:
RxNorm at 6 years. J Am Med Inform Assoc. v18 i4.
441-448.
Rink, B., Sanda Harabagiu, and Kirk Roberts, 2011.
Automatic extraction of relations between medical
concepts in clinical texts. Journal of the American
Medical Informatics Association 18.5: 594-600.
Rosario, B., Hearst, M. A., Ed. (2004). Classifying
Semantic Relations in Bioscience Texts
. In
Proceedings of the 42th Annual Conference of the
Association for Computational Linguistics.
Rudd, K. L., Johnson, M. G., Liesinger, J. T., & Grafft, C.
A, 2010. Automated detection of follow-up
appointments using text mining of discharge records.
International Journal for Quality in Health Care: 229-
235.
Salvadores, M., Alexander PR, Fergerson RW, Musen
MA, and Noy NF, 2012. Using SPARQL to Query
BioPortal Ontologies and Metadata. International
Semantic Web Conference. Boston US. LNCS 7650:
180-195.
Savona G. K, Masanz J. J., Ogren P. V., Zheng J., Sohn S.,
Kipper-Schuler K. C., Chute C, 2010. Mayo clinical
Text Analysis and Knowledge Extraction System
(cTAKES): architecture, component evaluation and
applications. J Am Med Inform Assoc. 2010 Sep-Oct;
17(5): 507–513. doi: 10.1136/jamia.2009.001560
Sibanda, T., et al. , 2006. Syntactically-informed semantic
category recognizer for discharge summaries. AMIA
annual symposium proceedings.
SNOMED-CT. "International Health Therminology
Standards Development Organisation." SNOMED-CT.
Retrieved 23.07, 2013, from
http://www.ihtsdo.org/snomed-ct/.
SPARQL 1.1 Query Language. W3C Recommendation 21
March 2013. http://www.w3.org/TR/sparql11-query/.
WHO - World Health Organization. 2004. International
Statistical Classification of Diseases and Health
Related Problems. G. W. H. Organization.
KDIR2014-InternationalConferenceonKnowledgeDiscoveryandInformationRetrieval
268
