
Semantic Enrichment of Relevant Feature Selection Methods
for Data Mining in Oncology
Adriana Da Silva Jacinto
1,2
, Ricardo Da Silva Santos
2,3
and José Maria Parente De Oliveira
2
1
Department of Computer Science, Faculty of Technology, São José dos Campos, Brazil
2
Department of Computer Science, Aeronautics Institute of Technology, São José dos Campos, Brazil
3
Integrated Center of Biochemistry Research, University of Mogi das Cruzes, Mogi das Cruzes, Brazil
1 STAGE OF THE RESEARCH
Medical field is one of the most important for
society, in which several researches are performed.
One of the main concerns of responsible
organizations is cancer, which is a disease in which
abnormal cells divide uncontrollably and invade
other tissues, forming a malignant tumor in the most
of the cases (Jemal, 2011; NCI, 2014).
In 2025, the number of new cases of the disease
in the world is estimated around 19.3 million (Ferlay
et al., 2012; Bray et al., 2013), i.e., the number of
cases of the disease increases and the discovery of
the cure seems remote.
Due to that situation, several medical databases
of patients related to cancer are provided for
research and analysis (NCI, 2014). The goal is to
search improvements on the prognosis, diagnosis,
prevention, treatment for the disease, the actions and
decision-making by public health managers.
Use of data mining is presented as an aid to the
analysis of those data, because it is the exploitation
and analysis of databases through a variety of
statistical techniques and machine learning
algorithms for the discovery of rules and relevant
patterns (Tan et al., 2005).
The success of data mining is subordinate to the
selection of the most relevant features (Lin et al.,
2006). However, the correct selection of features
depends on methods and is highly related to the
knowledge of domain experts, because the meanings
of some features are not easily understood by a
person who is not familiar with the application
domain, complicating the insight about which of
them are useful for data mining (Kuo et al., 2007).
Generally, oncology specialists are not available
to help a data miner full time. Moreover, the medical
field has several terms and peculiarities, which may
make hard the selection of the semantically most
relevant features. Additionally, the analysis of the
importance of the meaning of each feature in a data
set with thousands of genes as features, for example,
would require too long time of a human expert,
making it nonviable.
The high number of features in databases is
pointed out by several researchers (Hall, 2000;
Freitas, 2001; Inbarani et al., 2007; Tan et al., 2009;
Mansingh et al., 2011) as one of the crucial
problems in data mining. Over time, in order to
remedy this problem, various selection methods
were developed to improve the functionality of the
pre-processing of the data (Ammu e Preeja, 2013;
Chahkandi, 2013).
As seen in this work, those methods only employ
statistical techniques to select the features and do not
capture the semantics of them. As result, the more
semantically relevant features are excluded for not
having statistical significance, while irrelevant
features are selected due to their extensive statistical
contribution.
Thus, this project presents a proposal of
capturing of the semantic importance of each feature
by computational manner. The proposal enriches
the traditional methods of feature selection by using
of Natural Language Processing, the NCI ontology,
WordNet and medical documents.
A prototype of this approach was implemented
and tested with five data sets related to cancer
patients. The results show that the use of semantic
improves the pre – processing by selecting of the
most relevant semantic features.
2 OUTLINE OF OBJECTIVES
This work aims to:
Define an architecture for semantic enrichment
of feature selection filter methods, that
considers the semantics of the input features in
relation to predictable class;
Implement the architecture and apply it in
Oncology;
24
Da Silva Jacinto A., Da Silva Santos R. and Parente De Oliveira J..
Semantic Enrichment of Relevant Feature Selection Methods for Data Mining in Oncology.
Copyright
c
2014 SCITEPRESS (Science and Technology Publications, Lda.)

Evaluate whether the proposed approach
presents improvements to the process of data
mining.
3 RESEARCH PROBLEM
According to Lee (2005), the problem of feature
selection can be examined from different
viewpoints, and one of which is the search for the
most relevant features, i.e., the searching for those
feature that bring more meaning to the obtained data
mining model. However, generally, the feature
selection methods use statistical techniques and do
not consider the semantics of each attribute.
Due to the fact of the feature selection methods
do not consider the semantic of features the
following problems can arise: a) elimination of
semantically relevant features that do not show their
importance in statistical data analysis; b)
consideration of irrelevant or redundant features due
to their high level of statistical contribution.
This work consider irrelevant feature regarding
knowledge to be extracted is the one, if removed,
causes no impact on data mining model, and
redundant feature is one whose value is already
represented by another.
Wu et al. (2011) highlight that data mining is a
complicated activity for beginners in the area,
resulting in the specification of several parameter
settings or selection of features that are syntactically
accepted, but are not semantically reasonable.
Also true there is no mechanism that can help
users on the semantics of configurations, it means
that the user has difficulty when trying to identify
which features must be selected for a given data
mining task, even after having a suggested list of
features given by some selection method (Wu et al.,
2011). That occurs, as made by Microsoft Visual
Studio (Microsoft, 2014), because methods of
selection of features do not consider the semantics
between features and provide a list of selected
features by statistical means, whose refinement is
left to the user, can leave him confused.
Presented the context, the research problem of
this paper is enunciated as follow:
Current feature selection methods do not
consider the semantics of each feature, and the
result is the selection of irrelevant or redundant
data, which increase the computational cost of the
operation and the generation of data mining models
with low quality.
4 STATE OF THE ART
Since 1990, a variety of feature selection methods
have been implemented using different approaches
and techniques. Nevertheless, this study sought to
identify the core of the approach used on several
feature selection methods in order to group them by
similarities.
Considered methods are only those of the filter
type, it means that their implementation and
execution are independent of the mining algorithm.
Thus, the main approaches for feature selection in
the literature are based on Entropy, Consistency,
Matrix Resources, Rough Sets, Similarity and
Correlation.
Entropy and others (the probability calculus,
symmetric uncertainty, information gain, mutual
information and frequency values) quantify the
disorder among the elements of a data set, with basis
on the calculation of probability (Cover and
Thomas, 1991). When a data set is homogeneous,
lower is the entropy among its elements.
Feature selection methods consider that high
entropy of an input attribute relative to a predictable
class shows its relevance. Main idea is that
variations in the values of a feature generate a
greater degree of uncertainty concerning what
happens in the prediction feature (Hall, 2000).
Examples of feature selection methods that
employ entropy and similar are: Focus and Focus-2
(Almuallim and Dietterich, 1991, 1992);
Correlation-based Feature Selection (CFS) (Hall,
2000); Based Fast Correlation Filter (FCBF) (Yu
and Liu, 2003); Interact (Zhao and Liu, 2007);
Information Theoretic-based Interact (IT-IN) (Deisy
et al, 2010.); TRS + Focus2 (Teruya, 2008);
Information Gain Attribute Ranking (Cover and
Thomas, 1991). Other approach adopted by the
feature selection methods is the calculation of
consistency, which refers to coherency. Whether
there are two tuples with the same values of input
feature, they must have equal values for the
prediction feature. If it does not, set up an example
of inconsistency (Dash and Liu, 2003; Liu and
Setiono, 1996). Thus, the subset of input features
which displays the lowest level of inconsistency will
be chosen by the selection method.
Following feature selection methods use the
calculation of consistency to elect the most relevant
input features: Las Vegas Filter (LVF) and
Consistency Subset Evaluation (CSE) (Liu and
Setiono, 1998); Information Theoretic-based Interact
(IT-IN) (Deisy et al, 2010.); Focus and Focus-2
(Almuallim and Dietterich, 1991, 1992) and Interact
SemanticEnrichmentofRelevantFeatureSelectionMethodsforDataMininginOncology
25

(Zhao and Liu, 2007).
Proceeding the description of the approaches
found, there is the use of matrix resource (matrix
SVD Laplacian matrix), which consists of the
decomposition of a data matrix into singular values,
calculating the cosine between two columns or
covariance matrix. Each column of the matrix is
considered as a vector. The aim is to modify the
original data space, identifying input features that
can be disregarded, due to its low contribution to the
set of features, measured by calculating the
eigenvalues (Pearson, 1901).
Use of matrix resources is checked out on the
following features selection methods: Spectral
Feature Selection (Spec) (Zhao et al, 2007.);
Laplacian Score (He et al, 2005.); Principal
Components Analysis (PCA) (Pearson, 1901).
Whereas a data set is formed by different
vectors, these vectors can be compared, establishing
between them a measure, which is calculated with
basis on a metric. The metric can be the cosine of
the resulting angle, the Euclidean distance or
Manhattan distance between the vectors and some
variations. This approach is called similarity or
correlation, and vectors can be tuples or features
(Meira Jr. and Zaki, 2009).
Use of similarity and correlation happens in the
following selection methods: Minimum-
Redundancy-Maximum-Relevance (mrMr) (Peng et
al, 2005.); Redundancy Demoting (RD) (Osl et al,
2009.); FCBF (Yu and Liu, 2003); ReliefF ReliefF-1
and-2 (Rendell and Kira, 1992; Kononenko, 1994);
CFS (Hall, 2000); Network-based feature selection
approach (Netzer et al, 2012.); Redundancy Based
Filter (RBF) (Ammu and Preeja 2013).
Concluding the description of the approaches,
the Rough Sets theory conducts tests with all
possible subsets of input features by checking out
which one has better quality of approximation to the
original set (Pawlak, 1982, Hein and Kroenke,
2010).
Examples of feature selection methods
employing this approach are: Rough Sets Theory
(Hein and Kroenke, 2010; Pawlak, 1982); TRS +
Focus2 (Teruya, 2008); RSARSubsetEval (RSAR)
(Chouchoulas and Shen, 2001).
Note that some feature selection methods employ
more than one approach in order to obtain a more
relevant subset of features, refining the selection.
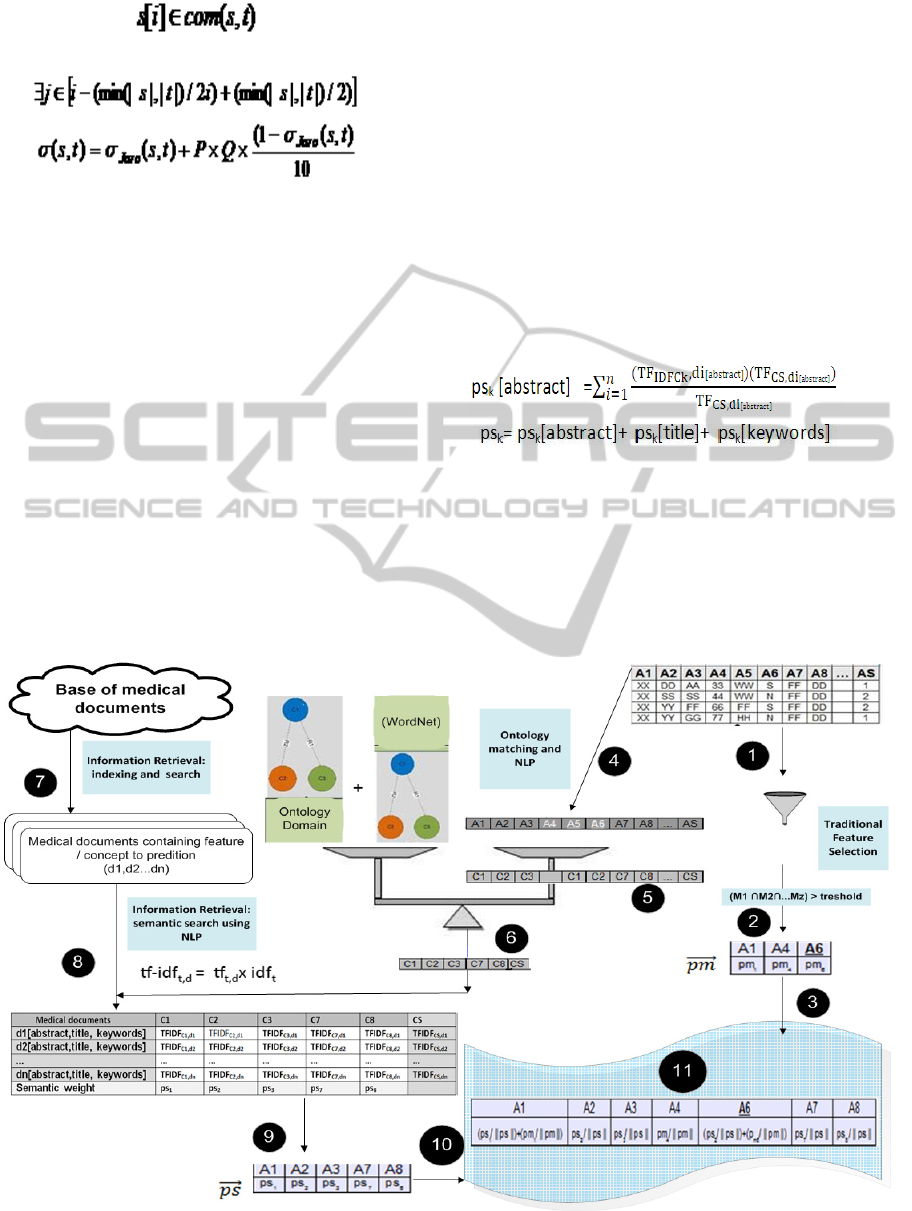
5 METHODOLOGY
Figure 1 presents the proposal of this work to enrich
semantically the feature selection methods.
Basically, this approach is divided into 11 steps as
follow.
1 – A data set with x features {A1, A2...,Ax}, y
tuples and a predictable class {AS} is the input of the
application.
2 – A combination of feature selection filter
methods chooses the most relevant features of the
data set, {M1, M2, …}. This choosing is based just
on the statistical analysis. The possible number of
combination is given by formula (1), where n is the
amount of available feature selection methods and p
is the used quantity of them.
(1)
3 – A subset of the original features is selected,
each one with respective statistical weight (pm). In
this step, it is possible to set up a threshold for the
feature to be accepted and at least one of the feature
selection methods must rank the features.
4 – From the data set, only the name of features
are taken. Those names are compared to an ontology
domain and to a lexical ontology. The used ontology
domain is National Cancer Institute Ontology (NCI,
2014) and the WordNet is a lexical ontology
(Fellbaum,1998; Miller, 1995). Lexical ontology is
used on Thesaurus or dictionaries to recognize
words.
5 – According to the relations between the
features and the predictable class, the names of
features are transformed on concepts from ontology
domain by the use of natural language processing
techniques. If some feature is not found on the
ontology domain, an automatic search for synonyms,
hyperonymy and other relations occurs on WordNet
(Fellbaum,1998; Miller, 1995).
This proposal performs an automatic
normalization procedure with the names of features,
which is the treatment of strings. This procedure
converts strings to lowercase, discards grammatical
accents, deletes blank spaces and hyphens,
withdrawal of numeric digits and punctuation. After
normalization of the strings, the comparison between
feature and an ontology concept is made by
calculating the similarity measure of words (Jaro,
1989; Jaro, 1995; Winkler, 1990) shown in formula
(6), and withdrawing Euzenat Shvaiko (2007).
(2)
(3)
IC3K2014-DoctoralConsortium
26
(4)
If only if
(5)
(6)
Letters s and t represent two strings to be
compared. Expression com(s,t) represents the
amount of characters that appears in the two strings,
but in a different order. Expression transp (s,t) refers
to the quantity of transpositions of characters
occurred. First calculating σ(s,t) refers to σ
Jaro
(s,t)
calculation. Second σ(s,t) calculation refers to the
measure of σJaro-Winkler (s,t), where P refers to the
size of the common prefix of two strings, and Q is a
constant.
6 – Just the features related to an equivalent
concept are pre – selected as semantically relevant.
Features related to a repeated concepts are removed
because this indicates redundancy. Initially, if a
features is not connected to a concept, it is
considered semantically irrelevant. Then a set of
important concepts, {C1, C2, C3, C7, C8,... CS}, is
selected and will be used on the next steps.
7 – From a base of medical documents, a set of
documents {d1, d2, d3… dn} that contains the
concept equivalent to predictable class (CS) are
recovered. The search looks into abstract, title and
keyword of the documents.
8 – Each concept came from the step 6 is
searching into documents came from step 7.
9 – Each concept k receives a semantic weight
(psk). This semantic weight is calculated by the
formula (8), which uses a modified and weighted
TF-IDF (Term Frequency Inverse Document
Frequency) (Salton e Buckley,1988). Final semantic
weight of each concept is the sum of semantic
weight into each field of the documents: abstract,
title and keyword.
(7)
(8)
10 – Each feature receives its respective
semantic weight, accordingly to related concept.
11 – Subsets came from steps 3 and 10 are faced.
One of three situations can occur:
If a feature is semantically and statistically
relevant, it receives the normalized sum of
weights (ps and pm);
Figure 1: Architecture of semantic enrichment of feature selection methods.
SemanticEnrichmentofRelevantFeatureSelectionMethodsforDataMininginOncology
27
If a feature is just statistically relevant, receives
the normalized statistical weight (pm/||pm||);
If a feature is just semantically relevant,
receives the normalized semantic weight
(ps/||ps||).
The output of this work is a subset of the most
relevant features.
To understand the formula (8), an example of
calculus of the semantic weight (psk[abstract]) for
the concept Ck was presented in formula (7),
considering the abstract of all selected documents.
Cited calculus has the follow stages:
TF-IDFck,di is calculated by the multiplication
of TFck,di by IDFck,di, where TFck,di is a
frequency of the concept Ck on document i;
and IDFck,di is the natural logarithm of the
number n of all selected documents divided by
the number of documents that contain the
concept Ck plus 1;
As the same way, the TF-IDFcs,di would be
calculated, but all selected documents contain
the concept CS related to the predictable
feature. So, just the TFcs,di is important to find
the weighted average psk[abstract];
Analogously, the weighted average for the
other fields of each document is calculated, in
this case psk [title] and psk[keywords].;
Final semantic weight of the concept Ck is the
sum of the three partial semantic weights.
Each subset of weights is considered a vector. So,
Euclidean Norm was used as presented in formulas
(9) and (10). It is used to normalize semantic weight
(ps) and statistical weight (pm).
(9)
(10)
6 EXPECTED OUTCOME
In theory, use of semantic enrichment feature
selection methods will bring benefits such as: a)
reduction of the required time to produce mining
models more coherent in the area of cancer and
tumors; b) facilitating of the construction of models
for data mining, since a data miner, without much
knowledge of the physician or genetic field, can
produce good mining models from the semantic
selection of features.
A prototype of the proposal was tested with five
data set related to patients with cancer or tumor as
follow.
Lymphograhy contains 18 features from 148
patients. Prediction feature is the diagnosis if a
patient is normal, has metastasis, has a malignant
lymphoma or a fibrosis.
Breast Cancer has 9 features and the goal is to
classify if there is risk of recurrence in patients who
have already received treatment for tumor.
Location of Primary Tumor presents 17
features of 339 patients. Prediction feature is to
know where primary tumor appeared.
Lymphograhy, Breast Cancer and Location of
Primary Tumor were obtained from the University
Medical Center, Institute of Oncology, Ljubljana,
Yugoslavia and available in http://archive.
ics.uci.edu/ml/. Thanks to M. Zwitter and M. Soklic
for providing this information.
Authorization for Hospitalization from
Brazilian Public Health System (AIH – SUS) - This
data set was obtained from the SIH files available on
site http://www2.datasus.gov.br/SIHD/, option
"Reduced the AIH" menu. It contains 30 attributes
related to hospitalization of 120325 patients with
brain tumors. Prediction feature is the period of stay
of patients, classifying it as short, medium, long or
very long.
Central Nervous System (CNS) is available at
http://www-genome.wi.mit.edu/mpr/CNS/. It con-
tains data of 7129 genes from 60 patients who had
cancer of the central nervous system and had
treatment. Prediction feature is to know if a patient
had survival or not, according to his genetic
characteristics.
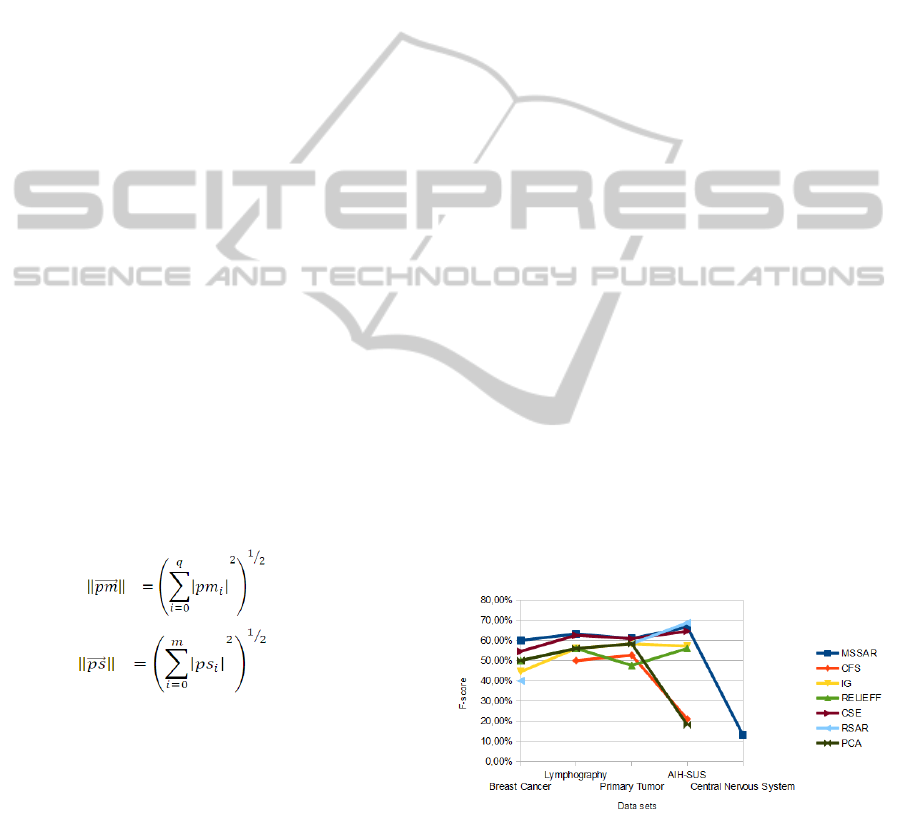
Figure 2: Comparative graph for each approach and data
set.
These five data sets were submitted to 6 approaches
of feature selection methods and the results were
compared with the literature concerning Oncology.
Literature came from different sources, from
separate source of the medical repository used to
retrieve documents in this research proposal.
IC3K2014-DoctoralConsortium
28

Features pointed out in the literature was considered
the gold pattern and F-score calculus was performed
for each approach. Figure 2 presents a comparative
graph.
F-score shows how much the selection of
features by an approach is close to literature. In
other words, how much a feature selection method is
according to human expert and semantic election.
The choosing of some approaches in specifics
data sets is very meaningless, according literature.
For example, this occurs on CFS with Breast Cancer
and on RSAR with Lymphograpy, because there is
no point for them.
For Central Nervous System data set, only the
proposal of this work, named MSSAR, had points.
MSSAR had points in all tested data sets. More
details concerning the tests are available at https://
drive.google.com/folderview?id=0B1CL3P-wkasBT
VBCZFB0UDFadncandusp=sharing.
REFERENCES
Almuallim, H.; Dietterich T. G. 1991. Learning with
Many Irrelevant Features. In: Proceedings of the 9th
National Conference on Artificial Intelligence,
Anaheim, CA, v. 2, pp. 547–552.
Almuallim, H.; Dietterich, T. G. 1992. Efficient algorithms
for identifying relevant features. In: Proceedings of the
Ninth Canadian Conference on Artificial Intelligence,
Vancouver, BC: Morgan Kaufmann. May 11-15, pp.
38-45.
Ammu, P. K.; Preeja, V. 2013. Review on Feature
Selection Techniques of DNA Microarray Data. In:
International Journal of Computer Applications 0975
– 8887 Volume 61– No.12, January 2013. pp. 39-44.
Bray, F.; Ren, J. S.; Masuyer, E.; Ferlay, J. Estimates of
global Cancer prevalence for 27 Sites in the Adult
Population in 2008. Int J Cancer. 2013 Mar 1; 132
(5):1133-45. doi:10.1002/ijc.27711. Epub 2012 Jul 26.
Chahkandi,Vahid; Yaghoobi, Mahdi; Veisi, Gelareh. 2013.
Feature Selection with Chaotic Hybrid Artificial Bee
ColonyAlgorithm based on Fuzzy CHABCF In:
Journal of Soft Computing and Applications. pp. 1-8
Chouchoulas, A.; Shen, Q. 2001. Rough set-aided keyword
reduction for text categorization. Applied Artificial
Intelligence: An International Journal. 159:843-873.
Cover, T. M.; Thomas, J. A. 1991. Elements of Information
Theory. Copyright © 1991 John Wiley and Sons, Inc.
Print ISBN 0-471-06259-6 Online ISBN 0-471-20061-
1. 563 p.
Dash, M.; Liu, H. 2003. Consistency-Based Search in
Feature Selection. Artificial Intelligence. 1511-2:155-
176, December, 2003.
Deisy, C., Baskar, S., Ramraj, N., Saravanan Koori, J., and
Jeevanandam, P. 2010.. A novel information theoretic-
interact algorithm (IT-IN) for feature selection using
three machine learning algorithms. Expert Systems
with Applications, 37(12), 7589-7597. Elsevier Ltd.
doi:10.1016/j.eswa.2010.04.084
Euzenat, Jérôme and Shvaiko, Pavel. 2007. Ontology
matching, Springer-Verlag, 978-3-540-49611-3.
Fellbaum, Christiane. 1998. WordNet: An Electronic
Lexical Database. Cambridge, MA: MIT Press.
Ferlay, J.; Soerjomataram, I.; Ervik, M. R.; Dikshit, S.;
Eser, C.; Mathers, M.; Rebelo, M.; Parkin, D.;
Forman, D.; Bray, F. GLOBOCAN 2012 v1.0, Cancer
Incidence and Mortality Worldwide: IARC Cancer
Base No. 11 [Internet]. Lyon, France: Inter-national
Agency for Research on Cancer; 2013. Available at:
http://globocan.iarc.fr, Accessed on June 2014.
Freitas, A. A. 2001. Understanding the Crucial Role of
Attribute Interaction in Data Mining. Artificial
Intelligence Review, (1991), 177-199.
Hall, M. A. 2000. A correlation-based feature selection for
discrete and numeric class machine learning.
ICML'00. In: Proceedings of the 17th International
Conference on Machine Learning. pp. 1157-1182.
He, X.; Cai, D.; Niyogi, P. 2005. Laplacian score for
feature selection. In: Y. Weiss, B. Scholkopf, and J.
Platt, editors, Advances in Neural Information
Processing Systems 18, Cambridge, MA, MIT Press.
Hein, N.; Kroenke, A. 2010 Escólios sobre a Teoria dos
Conjuntos Aproximados – Commentaries about the
Rough Sets Theory. In: Revista CIATEC – UPF, vol.2
1, pp. 13-20. doi: 10.5335/ciatec.v2i1.876 13.
Inbarani, H. H.; Thangavel, K.; Pethalakshmi, A. 2007.
Rough Set Based Feature Selection for Web Usage
Mining. In: International Conf. on Computational
Intelligence and Multimedia Applications. ICCIMA
2007, pp. 33-38. IEEE. doi:10.1109/ICCIMA.2007.356
Jaro, M. A. 1989. Advances in record linkage methodology
as applied to the 1985 census of Tampa Florida.
Journal of the American Statistical Association 84
(406): 414–20. doi:10.1080/01621459.1989.10478785.
Jaro, M. A. (1995). Probabilistic linkage of large public
health data file. Statistics in Medicine. 14 (5–7): 491–
8. doi:10.1002/sim.4780140510. PMID 7792443.
Jemal, A.; Bray, F.; Center, M.; Ferlay, J.; Ward, E.;
Forman., D. 2011. Global Cancer statistics. CA
Cancer Journal for Clinicians.; 61(2):69–90.
Kira, K.; Rendell, L. A. 1992. The Feature Selection
Problem: Traditional Methods and a New Algorithm,
In: Proceedings of 10th Conference on Artificial
Intelligence, Menlo Park, CA, pp. 129-136.
Kira, K.; Rendell, L. A.1992. A practical approach to
feature selection. In: Sleeman and P. Edwards,editors,
Proceedings of the 9th International Conference on
Machine Learning ICML-92, Morgan Kaufmann, pp.
249-256.
Kononenko, I. 1994. Estimating attributes: Analysis and
extension of RELIEFF. In: F. Bergadano and L. de
Raedt, editors, In: Proceedings of the European
Conference on Machine Learning, April 6-8, Catania,
Italy, Berlin: Springer-Verlag, pp. 171-182.
Kuo, Y-T.; Lonie, A.; Sonenberg, L. Domain Ontology
Driven Data Mining: A Medical Case Study.
Proceddings of 2007 ACM SIGKDD Workshop on
SemanticEnrichmentofRelevantFeatureSelectionMethodsforDataMininginOncology
29

Domain Driven Data Mining (DDDM2007); 2007.
Aug 12-14; San Jose, California, USA, pp.11-17.
Lee, Huei Diana. Seleção de Atributos Importantes para a
Extração de Conhecimento de Bases de Dados. Tese
de Doutorado. USP, 2005. 154p.
Liu, H.; Setiono, R. 1996. A Probabilistic Approach to
Feature Selection: a Filter Solution. In: Proceedings of
the 13th International Conference on Machine
Learning, Morgan Kaufmann. pp. 319–327
Liu, H.; Setiono, R.1998. Feature Selection for Large
Sized Databases. In Proceedings of the 4th World Con-
gress on Expert System, Morgan Kaufmann, pp. 68–75.
Mansingh, G.; Osei-Bryson, K.-M.; Reichgelt, H. 2011.
Using ontologies to facilitate post-processing of
association rules by domain experts. Information
Sciences, 1813, Elsevier Inc. pp. 419-434.
doi:10.1016/j.ins.2010.09.027.
Microsoft 2014. [Online]. Available at: <http://
msdn.microsoft.com/pt-br/library/ms175382.aspx>.
Accessed in 2014.
Miller, George A. 1995. WordNet: A Lexical Database for
English. Communications of the ACM Vol. 38, No.11:
39-41
National Cancer Institute (NCI) [Online]. Available at:
http://www.cancer.gov/ . Accessed in 2014.
Netzer, M.; Fang, X.; Handler, M.; Baumgartner, C. 2012.
A coupled two step network-based approach to
identify genes associated with breast cancer. Proc. 4th
Int. Conf. on Bioinformatics, Biocomputational Sys-
tems and Biotechnologies, (Biotechno, 2012), pp. 1-5.
Osl, M.; Dreiseitl, S.; Cerqueira, F.; Netzer, M.; Pfeifer,
B.; Baumgartner, C. 2009. Demoting redundant
features to improve the discriminatory ability in
cancer data. Journal of Biomedical Informatics,
424,Elsevier Inc. pp. 721-725. doi:10.1016/
j.jbi.2009.05.006
Pawlak, Z. 1982. Rough sets. In: International Journal of
Computer and Information Sciences, vol. 11, New
York, NY. n.º5, pp. 341-356, Plenum. http://roughsets.
home.pl/www/.
Pearson, K. 1901. On Lines and Planes of Closest Fit to
Systems of Points in Space. Philosophical Magazine 2
11: 559–572.
Peng, H.; Long, F.; Ding, C. 2005. Feature Selection
Based on Mutual Information: Criteria of Max-
Dependency, Max-Relevance, and Min-Redundancy.
IEEE Transactions on Pattern Analysis and Machine
Intelligence, 278: pp. 1226-1238.
Salton, G. and Buckley, C. 1988. Term-weighting
approaches in automatic text retrieval. Information
Processing and Management 24 (5): 513–523.
doi:10.1016/0306-4573(88)90021-0..
Tan, K. C.; Teoh, E. J.; Yu, Q.; Goh, K. C. 2009. A hybrid
evolutionary algorithm for attribute selection in data
mining. Expert Systems with Applications, 364, pp.
8616-8630. doi:10.1016/j.eswa.2008.10.013
Tan, P.-N.; Steinbach, M.; Kumar, V. 2005. Introduction to
Data Mining, 1st Edition. Addison-Wesley Longman
Publishing Co., Inc., Boston, MA, USA.
Teruya, Anderson. 2008 Uma metodologia para seleção de
atributos no processo de extração de conhecimento de
base de dados baseada em teoria de rough sets.
Dissertação de Mestrado. Universidade Federal Mato
Grosso do Sul, 86p.
Winkler, W. E. 1990. String Comparator Metrics and
Enhanced Decision Rules in the Fellegi-Sunter Model
of Record Linkage. In: Proceedings of the Section on
Survey Research Methods (American Statistical
Association): 354–359.
Wu, C.-A., Lin, W.-Y., Jiang, C.-L., and Wu, C.-C. 2011.
Toward intelligent data warehouse mining: An
ontology-integrated approach for multi-dimensional
association mining. Expert Systems with Applications,
38(9), 11011-11023. Elsevier Ltd. doi:10.1016/j.eswa.
2011.02.144.
Yu, L.; Liu, H. 2003. Feature selection for high-
dimensional data: A fast correlation-based filter
solution. In: T. Fawcett and N. Mishra, editors,
Proceedings of the 20th International Conference on
Machine Learning ICML-03, August 21-24, Washing-
ton, D.C., 2003. Morgan Kaufmann, pp. 856-863.
Zaki, M.; Meira Jr, W.2009 Fundamentals of Data Mining
Algorithms, Cambridge University Press in press.
555p. Available at: http://www.dcc.ufmg.br/mining
algorithms/
Zhao, Z.; Liu, H.2007. Searching for Interacting Features.
In: Proceedings of the 20th International Joint
Conference on AI IJCAI, January 2007.
Zhao, Z.; Liu, H. 2007. Spectral feature selection for
supervised and unsupervised learning. In International
Conference on Machine Learning ICML, 2007.
IC3K2014-DoctoralConsortium
30
