
A Nonlinear Mixture Model based Unsupervised Variable Selection
in Genomics and Proteomics
Ivica Kopriva
Ruđer Bošković Institute, Bijenička cesta 54, 10000 Zagreb, Croatia
Keywords: Variable Selection, Nonlinear Mixture Models, Explicit Feature Maps, Sparse Component Analysis.
Abstract: Typical scenarios occurring in genomics and proteomics involve small number of samples and large number
of variables. Thus, variable selection is necessary for creating disease prediction models robust to
overfitting. We propose an unsupervised variable selection method based on sparseness constrained
decomposition of a sample. Decomposition is based on nonlinear mixture model comprised of test sample
and a reference sample representing negative (healthy) class. Geometry of the model enables automatic
selection of component comprised of disease related variables. Proposed unsupervised variable selection
method is compared with 3 supervised and 1 unsupervised variable selection methods on two-class
problems using 3 genomic and 2 proteomic data sets. Obtained results suggest that proposed method could
perform better than supervised methods on unseen data of the same cancer type.
1 INTRODUCTION
Microarray gene expression profiling technology
(Alon et al., 1999; Shipp et al., 2002; Singh et al.,
2002) and mass spectrometry (Mischak et al., 2009;
Petricoin et al., 2002a; Petricoin et al., 2002b) are
key technologies used, respectively, to monitor gene
and protein expressions. Data from these types of
experiments present a "large p, small n" problem: a
very large number of variables (genes or m/z ratios)
relative to the number of samples (gene or protein
expressions). Under such circumstances learned
diagnostic (prediction) models are very likely to
overfitt; that is not to generalize well to new data
from the same cancer type even though performance
on the training set was good (Statnikov et al.,
2005a; Guyon et al., 2002). To improve diagnostic
performance of the predictors as well as to gain
better understanding of the underlying process that
generated data a variable selection is necessary
(Guyon et al., 2002; Statnikov et al., 2005a); that is
to select small number of genes or m/z ratios that
discriminate well between healthy and cancer
patients. Thereby, the goal is to select a subset of
variables that together have good predicting power,
rather than ranking them according to their
individual predicting powers. Methods for this
problem are grouped into wrappers, filters and
embedded methods (Guyon et al., 2002; Kohavi and
John, 1997; Lazar et al., 2012). Filters select subset
of variables as a pre-processing step independently
of the predictor. Wrappers use chosen learning
machine to score subsets of variables. Embedded
methods select variables together with the training of
predictor. Depending on whether diagnoses
information (class label) is used we distinguish
supervised (Peng et al., 2005) and unsupervised
methods in variable selection. Unsupervised
approaches are important for discovering novel
biological mechanisms and analyzing large datasets
for which little prior knowledge is available.
Because of no use of diagnoses information it is
expected that diagnostic models trained on variables
selected by unsupervised approaches generalize
better on unseen data from the same cancer type.
Unsupervised approaches can be further divided
into: clustering methods (Ben-Dor et al., 1999),
model-based methods (Lazzeroni and Owen, 2002),
and projection methods. A disadvantage of
clustering methods is that genes are partitioned into
mutually exclusive clusters, whereas in reality a
gene or experiment may be a part of several
biological processes. Model-based approaches first
generate a model that explains interaction among
biological entities participating in genetic regulatory
networks and then train the parameters of the model
on expression datasets. The challenge of model-
85
Kopriva I..
A Nonlinear Mixture Model based Unsupervised Variable Selection in Genomics and Proteomics.
DOI: 10.5220/0005161700850092
In Proceedings of the International Conference on Bioinformatics Models, Methods and Algorithms (BIOINFORMATICS-2015), pages 85-92
ISBN: 978-989-758-070-3
Copyright
c
2015 SCITEPRESS (Science and Technology Publications, Lda.)

based approaches may be the lack of sufficient data
to train the parameters. Projection methods
decompose dataset into components that have
desired property. Since variables in components
obtained by decomposition are latent or hidden these
methods are also known under the name latent
variable analysis. The most often used projection
methods are: principal component analysis (PCA),
independent component analysis (ICA), sparse
component analysis (SCA) and nonnegative matrix
factorization (NMF). PCA (Alter et al., 2000)
decomposes experimental data into uncorrelated
components. In contrast to PCA, ICA decomposes
input dataset into statistically independent
components (Hyvärinen et al., 2001). That yields
biologically relevant components that are
characterized by the functional annotation of genes
that are predominant within the component (Lee and
Batzoglou, 2003). SCA-based decomposition yields
biologically relevant components that are composed
of small number of genes (variables), i.e. they are
sparse (Schachtner et al., 2008; Stadtlthanner et al.,
2008; Gao and Church, 2005; Kim and Park, 2007).
NMF (Cichocki et al., 2010) decomposes dataset
into nonnegative components (Brunet et al., 2004)
that in addition can be sparse (Gao and Church,
2005; Kim and Park, 2007; Kopriva and Filipović,
2011). Unsupervised decomposition methods for
variable selection, the representatives of which are
referenced above, have the following limitations: (i)
they are based on a linear mixture model (Girolami
and Breitlling, 2004) representing dataset as
weighted linear superposition of components. The
exception is (Lee and Batzoglou, 2003) where, in
addition to linear, nonlinear mixture model is used to
represent gene expressions and that has been
motivated by the fact that interactions within gene
regulatory networks can be nonlinear (Yuh et al.,
1998); (ii) the whole dataset is used for
decomposition yielding only one component with
cancer related variables. This component can be
used for biomarker identification studies but it does
not suffice to learn diagnostic model.
Here we propose wrapper-like variable selection
method. It performs unsupervised variable selection
by individually decomposing each sample into
sparse components. Thereby, decomposition is based
on nonlinear mixture model comprised of considered
sample and a reference sample representing negative
(healthy) class. The model is nonlinear
generalization of the linear mixture model with a
reference sample presented in (Kopriva and
Filipović, 2011). Nonlinear mapping is performed
across sample dimension yielding possibly linear
model with preserved number of variables and
"increased" number of samples. Sparseness
constrained decomposition is performed in mapped
space, whereas selection of component with cancer
related variables is performed automatically (without
using diagnoses information). Afterwards, variables
in cancer related components are ranked by their
variance. This yields index set that is used to access
true variables in the original input space of samples.
They are used to learn diagnostic models by cross-
validating two-class support vector machine (SVM)
classifier (Vapnik, 1998). To make our results
reproducible Gene Expression Model Selector
(GEMS) software system has been used for cross-
validation and learning of SVM-based diagnostic
models. The system is available online at:
http://www.gems-system.org/. It uses the LibSVM
team (Chang and Lin, 2003) based implementation
of the SVM algorithms. The GEM implements two-
loops based system known as nested stratified cross-
validation (Statnikov et al., 2005a; Statnikov et al.,
2005b) that avoids overfitting. It has also been found
that diagnostic models produced by GEMS perform
well in independent samples and that GEMS-based
cross-validation performance estimates approximate
well the error obtained by the independent validation
(Statnikov et al., 2005b). Hence, it is believed that
performance estimate of proposed approach to
variable selection is trustworthy. Proposed approach
is compared with three state-of-the-art supervised
(Brown 2009; Aliferis et al., 2010) and one
unsupervised (Kopriva and Filipović, 2011) variable
selection method on three well-known cancer types
in genomics: colon cancer (Alon et al., 1999),
diffuse large b-cell lymphomas and follicular
lymphomas (Shipp et al., 2002) and prostate cancer
(Singh et al., 2002), and two well-known cancer
types in proteomics: ovarian cancer (Petricoin et al.,
2002a) and prostate cancer (Petricoin et al., 2002b).
Proposed method yields comparable accuracy
with slightly more variables than supervised
methods and it outperforms its linear counterpart
(Kopriva and Filipović, 2011).
The rest of the paper is organized as follows.
Proposed approach to variable selection is described
in section 2. Results of comparative performance
analysis are presented in section 3. Discussion and
conclusions are proposed in section 4.
2 METHODS
A sample recorded by microarray or mass
spectrometer contains components imprinted by
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
86

several interfering sources. As an example in
(Decramer et al., 2008) it is described how different
organs imprint their substances (components) into a
urine sample. These substances (components) can be
generated during disease progression and their
identification may be beneficial for early diagnoses
of disease (Mischak et al., 2009). That, however, is
complicated by the fact that component of interest
may be "buried" within a sample. Unsupervised
decomposition methods briefly elaborated
previously presume most often that sample is linear
superposition of components. This section presents
sparseness constrained unsupervised decomposition
method for variable selection using novel type of
nonlinear mixture model of a sample. The mixture
model is comprised of considered sample and a
reference sample that represents negative (healthy)
class. The model is nonlinear generalization of the
linear mixture model with a reference sample that
was presented in (Kopriva and Filipović, 2011).
2.1 Linear Mixture Model
Let us assume that N samples (gene or protein
expressions) are stored in rows of data matrix
NK
R
X , whereas each sample is further
comprised of
K variables (genes or m/z ratios). We
also assume that N samples have diagnoses (label):
1
,1,1
K
nn
Ry
x , n=1,...,N, where 1 stands for
positive (cancer) and -1 stands for negative (healthy)
sample. Matrix factorization methods such as PCA,
ICA, SCA and/or NMF assume linear mixture
model. For this purpose data matrix X is modelled as
a product of two matrices:
XAS
(1)
where
0
NM
R
A
,
M
K
R
S
and M stands for an
unknown number of components imprinted in
samples. Each component is represented by a row
vector of matrix S, that is:
1
K
m
R
s
, m=1,...,M.
Column vectors of matrix A:
1N
m
R
a
, m=1,...,M ,
represent concentration profiles of the corresponding
components. To infer component comprised of
disease relevant variables label information is used
by methods such as (Schachtner et al., 2008;
Liebermeister, 2002; Lee and Batzoglou, 2003).
Extracted component is further analyzed by
clustering to determine biological relevance and
extract biomarkers but it does not suffice to learn
diagnostic models. To address this limitation a linear
mixture model with a reference sample was
proposed in (Kopriva and Filipović, 2011):
1,...,
ref
nn
n
nN
x
AS
x
(2)
where
2
0
M
n
R
A
and
M
K
n
R
S
respectively
represent sample dependent matrices of
concentration profiles and components the number
of which,
M, was assumed to be the same for all the
samples. x
ref
stood for a reference sample that
represented either positive or negative class. Herein,
we assume that x
ref
represents negative (healthy)
class. It can be selected by an expert or, as it was the
case herein, can be obtained by averaging all the
samples belonging to negative class. As opposed to
linear mixture model (1), the linear mixture model
(2) has greater flexibility because the model is
sample adaptive. That addresses issue of biological
diversity, because even samples within the same
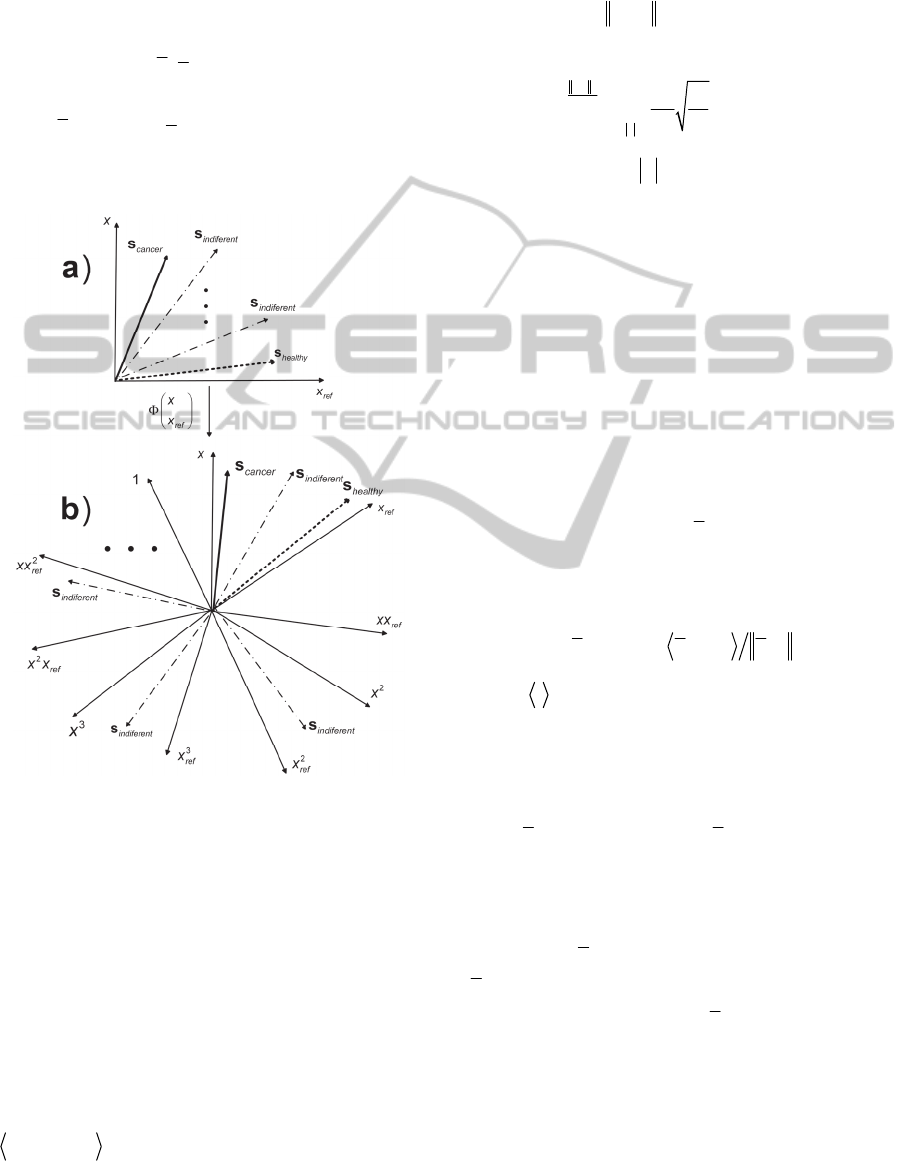
group are different. Geometry of the mixture model
(2) enables to automatically select component with
cancer relevant variables. Provided that x
ref
represents negative group component with cancer
relevant variables, s
cancer
, is the one associated with
the mixing vector that closes the largest angle with
the axis defined by the x
ref
sample. Component
comprised of variables related to healthy state,
s
healthy
, is the one associated with the mixing vector
that closes the smallest angle with the axis defined
by the x
ref
sample. The rest of the M-2 components
are comprised of differentially not expressed
(indifferent) variables. That is illustrated in Figure
1a.
2.2 Nonlinear Mixture Model
As pointed out in (Lee and and Batzoglou, 2003;
Yuh et al., 1998) interactions among components in
biological samples do not have to be linear only.
Thus, nonlinear mixture model would be more
general description. Nonlinear generalization of the
linear model (2) is given with:
,
;
1,..., ; 1,...,
ref k
nkn
nk
x
f
nNkK
x
s
(3)
where
2
:
n
M
n
f
RR
is an unknown sample
dependent nonlinear function that maps
M
n
-
dimensional vector of variables
1
;
n
M
kn
R
s to 2-
dimensional observation vector. Thereby, first
element of the observation vector belongs to the
reference sample and second element to the test
sample. Herein, we assume that reference sample
represents negative (healthy) class. It can be selected
by an expert or, as it was the case herein, can be
ANonlinearMixtureModelbasedUnsupervisedVariableSelectioninGenomicsandProteomics
87
obtained by averaging all the samples belonging to
negative class. Note that unlike the linear
counterpart (2) the nonlinear model (3) assumes that
number of components contained in the sample,
M
n
,
is also sample dependent. We map (3) explicitly:
,
;
1,...,
ref k
nkn
nk
x
kK
x
As
(4)
where
0
n
D
M
n
R
A
,
1
;
n
M
kn
R
s and
2
:
D
RR
is
nonlinear mapping that increases original number of
samples from 2 to
D>2.
Figure 1: Geometry of the mixture model with a reference
sample: a) linear model (2); b) nonlinear model (4).
The algebraic structure of the mapping is of the
form:
12
12
12
,
,
,0
12
s.t. 0
T
d
ref k
qq
qq refknk
qq
nk
x
cc x x
x
qq d
(5)
where
d is order of the mapping. Coefficients are
mapping dependent. We prefer mappings that induce
reproducible kernel Hilbert space (RKHS) of
functions and are, therefore, associated with the
kernel function through
kernel trick:, where
,
H
xy
denotes the inner product in RKHS
H
induced by kernel
. It enables us to construct
explicit mapping
by factorizing the kernel
function. We have chosen the Gaussian kernel:
2
2
,exp /xy x y
for which
factorization yields:
2
2
0
12
e1,...,
!
nk
r
d
nk nk
r
rr
kK
x
α
α
xx
α
(6)
such that
2
0
N
α
,
12
α
,
12
!!!
α
and
12
,nk ref k nk
x
x
α
x
. d is an order of approximation that is
data dependent and has to be determined by cross-
validation. Regarding
we have found that when
data are scaled to [-1, 1] interval,
can be
approximately set to 1. Dimension
D of mapping
induced space depends on order of the mapping d
through:
D=(d+2)(d+1)/2. We can write (5) as:
,
11,2 2 3
...
ref k
ref k nk
nk
x
cx cx
x
eee
(7)
where
D
j
Re
, j=1,...,D, are unit vectors that form
the orthonormal basis in
D
R
. Thus, cosines of the
angles that column vectors
;0
D
mn
R
a
, m=1, ..., M
n
,
in model (4), close with the axis defined by a
reference sample x
ref
in mapped space are obtained
as:
;;2;
cos , ,
mn ref mn mn
ax ae a
(8)
where
,
stands for inner product. When sample
x
ref
represents healthy class component comprised of
cancer relevant variables is associated with the
column vector that closes maximal angle with
respect to axis defined by a reference sample, that is:
;;
arg min cos ,
cancer n m n ref
m
sax
(9)
That is illustrated in Figure 1b. When each sample is
decomposed according to (4) components comprised
of cancer relevant variables (9) are stored row-wise
in a matrix
NK
cancer
R
S
. Variables (columns of
cancer
S
) are then ranked by their variance across the
sample dimension yielding
ranked N K
cancer
R
S
. Let us
denote by
I a corresponding index set. Variables
ranked in the original space of samples are obtained
by indexing each sample by
I, that is:
ranked
nn
xx
(I),
n=1,..., N. Samples with ranked variables form rows
of the matrix
ranked N K
R
X
that, when paired with
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
88

the vector of labels
y
, is used to learn SVM-based
diagnostic models.
2.3 Sparse Component Analysis
Decomposition of the linear mixture model (4) is
performed enforcing sparseness of the components
;mn
s
, m=1, .., M
n
. Sparseness constraint is in
microarray data analysis justified by biological
reasons (Stadtlthanner et al., 2008, Gao and Church,
2005). That is, sparse components are comprised of
few dominantly expressed variables and that can be
good indicator of a disease. In relation to a mixture
model with a reference sample (3)/(4) sparseness
constraint implies that variable is dominantly
expressed in: (
i) cancer related component and few
components comprised of differentially not
expressed variables; (
ii) few components comprised
of differentially not expressed variables; or (
iii) few
components comprised of differentially not
expressed variables and healthy class related
component.
Method used to solve, in principle,
underdetermined blind source separation problem
(4) estimates mixing matrix
n
A
first by using the
algorithm (Gillis and Vavanis, 2012) with a
MATAB code available at: https://sites.google.com/
site/nicolasgillis/publications. The important
characteristic of the method is that there are no free
parameters to be tuned or defined
a priori. The
unknown number of components
M
n
is also
estimated automatically and is limited above by D.
Thus, by cross-validating d we implicitly cross-
validate
M
n
as well. After
n
A
is estimated the
n
S
is
estimated by minimizing sparseness constrained cost
function:
2
n
1
1
ˆˆ
min
2
n
ref
nn n
n
F
S
x
SAS S
x
(10)
where the hat sign denotes an estimate of the true
(but unknown) quantity, is regularization
parameter and
1
n
S denotes
1
-norm of
n
S
. We
have used the iterative shrinkage thresholding (IST)
type of method (Beck and Teboulle, 2009) with a
MATLAB code at: http://ie.technion.ac.il/
Home/Users/ becka.html. A sparsity of the solution
is controlled by the parameter . There is a maximal
value of (denoted by
max
here) above which the
solution of the problem (10) is equal to zero. Thus,
in the experiments reported in section 3 the value of
has been selected by cross-validation with respect
to
max
. Proposed variable selection algorithm is
outlined in Table 1. Please note that by setting d=1
in (5)/(6) and by ignoring the first term we actually
perform decomposition of a linear mixture model
proposed in (Kopriva and Filipović, 2011).
Table 1: A nonlinear mixture model with a reference-
based algorithm for variable selection.
Inputs:
NK
R
X
data matrix with N rows
representing samples (gene or protein expressions) and
K columns representing variables (genes or m/z ratios);
1
1,1
N
n
n
y
labels or diagnoses;
1
K
ref
R
x
reference sample representing negative (healthy) group.
Scale the data matrix X such that -1x
nk
1, n=1,...,N
and k=1,...,K.
Nested stratified cross-validation.
Loop 1: order of nonlinear mapping in (5)/(6): d{1, 2,
3, 4, 5};
Loop 2: regularization constant in (10): {0.05, 0.1,
0.2, 0.3, 0.4, 0.5}
max
.
1. x
n
=X(n,:), n=1,...,N form a nonlinear mixture
model according to (2).
2. Perform variable-wise nonlinear mapping of (2) by
mapping (6) with =1 with chosen d.
3. According to (4) use separable NMF algorithm
(Gillis and Vavanis, 2012) to estimate mixing matrix
n
A
and IST algorithm (Beck and Teboulle, 2009) to
estimate
n
S
with chosen .
4. According to (9) select cancer related component
;
(,:)
cancer cancer n
n Ss
.
5. Rank selected variables in
cancer
S
by their variance to
obtain index set I.
6. Obtain selected variables in the original input space
as: X
ranked
=X(:,I).
7. Use (X
ranked
, y) to perform cross-validation with
optimal parameters of the SVM classifier with
polynomial and Gaussian kernels. Use normalization of
variables to [0, 1] interval.
End of loop 2.
End of loop 1.
8. Select diagnostic model with the highest accuracy.
ANonlinearMixtureModelbasedUnsupervisedVariableSelectioninGenomicsandProteomics
89

3 RESULTS
Proposed approach is compared against state-of-the-
art supervised variable selection methods: maximum
mutual information minimal redundancy (MIMR)
method (Brown, 2009) and HITTON_PC and
HITTON_MB (Aliferis et al., 2010) methods. We
also report results for linear counterpart of proposed
method (Kopriva and Filipović, 2011). Gene
Expression Model Selector (GEMS) software system
(Statnikov
et al., 2005b), has been used for cross-
validation and learning of SVM-based diagnostic
models with polynomial and Gaussian kernels the
parameters of which were optimized in cross-
validation loop as well. The system is available
online at: http://www.gems-system.org/. HITON_PC
and HITON_MB algorithms are implemented in
GEMS software system while implementation of the
MIMR algorithm is available at MATLAB File
Exchange. Methods were compared on three cancer
types in genomics: colon cancer (Alon et al
., 1999),
diffuse large b-cell lymphoma and follicular
lymphomas (DLBCL/FL) (Shipp et al., 2002) and
prostate cancer (Singh et al., 2002) and two cancer
types in proteomics: ovarian cancer (Petricoin et al.,
2002a) and prostate cancer (Petricoin et al., 2002b).
The five datasets are described in Table 2. For each
dataset we report the best result achieved by one of
these supervised methods. The results obtained by
10-fold cross-validation are reported in Table 3. Due
to the lack of space we do not report details on
parameters of the SVM classifiers. For each of five
datasets proposed method achieves result that is
worse than but comparable with the result of state-
of-the-art supervised algorithm and much better than
its linear unsupervised counterpart. Since reported
results are achieved with small number of variables
the probability of overfitting is reduced. Thus, it is
reasonable to expect that performance on unseen
data of the same cancer type by proposed
unsupervised method will be better than the one
achieved with supervised algorithms.
Colon cancer data are available at: http://genomic-
pubs.princeton.edu/oncology/affydata/index.html.
Prostate cancer and DLBCL/FL genomic data are
available at: http://www.gems-system.org/. Ovarian
and prostate cancer proteomic data (mass spectra)
are available at: http://home.ccr.cancer.gov/
ncifdaproteomics/ppatterns.asp. To comply with
principle of reproducible research software that
implements steps 1 to 6 of the proposed algorithm,
datasets used and results presented in Table 3 are
available at: http://www.lair.irb.hr/ikopriva/Data/
HRZZ/data/BIOINFORMATICS_2015.zip
Table 2: Cancer human gene and protein expression
datasets used in comparative performance analysis.
Dataset
Number of samples
(cancer/normal)
Number of
variables
Reference
1. Prostate
cancer
52 /50 10509
Singh et al.,
2002
2. Colon
cancer
40/22 2000
Alon et al.,
1999
3. DLBCL/FL 58/19 5469
Shipp et al.,
2002
4. Ovarian
cancer
100/100 15152
Petricoin et
al., 2002a
5. Prostate
cancer
69/63 15154
Petricoin et
al., 2002b
Table 3: Classification accuracy and number of selected
variables.
Dataset
Proposed
method
Supervised
method
(Kopriva,
Filipović,
2011)
1. Prostate
cancer
91.27% / 38
genes (d=2,
=0.4).
MIMR: 98.09% /
10 genes.
94.27% / 477
genes.
2. Colon
cancer
91.91% / 24
genes (d=5,
=0.1).
HITON_MB:
93.33% 4 genes.
90.48% / 30
genes, =0.05.
3. DLBCL/FL
96.25% / 14
genes (d=2,
=0.2).
HITON_PC:
100% / 6 genes.
98.57% / 169
genes, =0.01.
4. Ovarian
cancer
93% / 7 m/z
lines (d=4,
[0.4, 0.7]).
HITON_PC:
99.5% / 7 m/z
lines.
82% / 25 m/z
lines, =0.2.
5. Prostate
cancer
94.06% / 14
m/z lines
(d=4, =0.2).
MIMR: 100% /
10 m/z lines
94.01% / 85
m/z
lines,=0.2.
d denotes order of nonlinear mapping (6) and
denotes regularization parameter in (10).
4 CONCLUSIONS
Because it requires little prior knowledge
unsupervised decomposition of set of samples into
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
90

additive mixture of components is of particular
importance in addressing overfitting problem.
However, contemporary unsupervised
decomposition methods require label (diagnoses)
information to select component with cancer
relevant variables. Such component is useful for
biomarker identification studies but it does not
suffice to learn diagnostic model. In addition to that,
most of existing unsupervised decomposition
methods assume linear additive mixture model of a
sample. Herein, we have proposed an approach for
variable selection by decomposing each sample
individually into sparse components according to
nonlinear mixture model of a sample, whereas
decomposition is performed with respect to a
reference sample that represents negative (healthy)
class. This enables to select cancer related
components automatically and use them for either
biomarker identification studies or learning
diagnostic models. It is conjectured that outlined
properties of proposed approach to variable selection
enabled competitive diagnostic accuracy with small
number of variables on cancer related human gene
and protein expression datasets. While proposed
approach to variable selection is developed for
binary (two-class) problems its extension for multi-
category classification problems is aimed for the
future work.
ACKNOWLEDGEMENTS
This work has been supported through grant
9.01/232 funded by the Croatian Science
Foundation.
REFERENCES
Aliferis, C. F., et al. (2010a). Local Causal and Markov
Blanket Induction for Causal Discovery and Feature
Selection for Classification - Part I: Algorithms and
Empirical Evaluation. J. Mach. Learn. Res., 11, 171-
234.
Aliferis, C. F., et al. (2010b). Local Causal and Markov
Blanket Induction for Causal Discovery and Feature
Selection for Classification - Part II: Analysis and
Extensions. J. Mach. Learn. Res., 11, 235-284.
Alon, U., et al. (1999). Broad patterns of gene expression
revealed by clustering analysis of tumor and normal
colon tissues probed by oligonucleotide arrays. Proc.
Natl. Acad. Sci. USA, 96, 6745-6750.
Alter, O., Brown, P. O., and Botstein, D. (2000). Singular
value decomposition for genome-wide expression data
processing and modeling. Proc. Natl. Acad. Sci. USA,
97, 10101-10106.
Aronszajn, N. (1950). The theory of reproducing kernels.
Trans. of the Amer. Math. Soc., 68, 337-404.
Beck, A. and Teboulle, M. (2009). A fast iterative
shrinkage-thresholding algorithm for linear inverse
problems. SIAM J. on Imag. Sci., 2, 183-202.
Ben-Dor, A., Shamir, R., and Yakhini, Z. (1999).
Clustering gene expression patterns. J. Comp. Biol., 6,
281-297.
Brown, G. (2009). A New Perspective for Information
Theoretic Feature Selection. J. Mach. Learn. Res., 5,
49-56.
Brunet, J. P., et al. (2004). Metagenes and molecular
pattern discovery using matrix factorization. Proc.
Natl. Acad. Sci. USA, 101, 4164-4169.
Chang, C. C., and Lin, C. J. (2003). LIBSVM: a library for
support vector machines.
Cichocki, A., et al. (2010). Nonnegative Matrix and
Tensor Factorizations. John Wiley, Chichester.
Decramer, S., et al. (2008). Urine in clinical proteomics.
Mol Cell Proteomics, 7, 1850-1862.
Dudoit, S., Fridlyand, J., and Speed, T. P. (2002).
Comparison of Discrimination Methods for the
Classification of Tumors Using Gene Expression Data.
J. of the Amer. Stat. Assoc., 97, 77-87.
Gao, Y., and Church, G. (2005). Improving molecular
cancer class discovery through sparse non-negative
matrix factorization. Bioinformatics, 21, 3970-3975.
Gillis, N., and Vavanis, S. A. (2012). Fast and Robust
Recursive Algorithms for Separable Nonnegative
Matrix Factorization, arXiv , v2.
Girolami, M., and Breitling, R. (2004). Biologically valid
linear factor models of gene expression.
Bioinformatics, 20, 3021-3033.
Gribonval, R., and Zibulevsky, M. (2010). Sparse
component analysis. In Jutten, C., and Comon, P.
(eds.), Handbook of Blind Source Separation,
Elsevier, pp. 367-420.
Guyon, I., et al. (2002). Gene selection for cancer
classification using support vector machines. Machine
Learning, 46, 389-422.
Guyon, I., Elisseeff, A. (2002). An introduction to variable
and feature selection. J. of Machine Learning Res., 3,
1157-1182.
Harmeling, S., Ziehe, A., and Kawanabe, M. (2003).
Kernel-Based Nonlinear Blind Source Separation,
Neural Comput., 15, 1089-1124.
Hyvärinen A., Karhunen J., and Oja E. (2001).
Independent Component Analysis. John Wiley & Sons,
New York.
Hoyer, P. O. (2004). Non-negative matrix factorization
with sparseness constraints, J. Mach. Learn. Res., 5,
1457-1469.
Jutten, C., Babaie-Zadeh, M., and Karhunen, J. (2010).
Nonlinear mixtures. In Jutten, C., and Comon, P.
(eds.), Handbook of Blind Source Separation,
Elsevier, pp. 549-592.
Kim, H., and Park, H. (2007). Sparse non-negative matrix
factorizations via alternating non-negativity
constrained least squares for microarray data analysis.
ANonlinearMixtureModelbasedUnsupervisedVariableSelectioninGenomicsandProteomics
91

Bioinformatics, 23, 1495-1502.
Kohavi, R., and John, G. (1997). Wrappers for feature
selection. Artificial Intel., 97, 273-324.
Kopriva, I., and Filipović, M. (2011). A mixture model
with a reference-based automatic selection of
components for disease classification from protein
and/or gene expression levels. BMC Bioinformatics,
12, 496.
Kruskal, W., and Wallis, W. A. (1952). Use of ranks in
one-criterion variance analysis. J. of the Am. Stat.
Assoc., 47: 583–621.
Lazar, C., et al. (2012). A survey on filter techniques for
feature selection in gene expression microarray
analysis. IEEE Tr. Comp. Biol. and Bioinf., 9, 1106-
1119.
Lazzeroni, L., and Owen, A. (2002). Plaid models for gene
expression data. Statistica Sinica, 12, 61-86.
Lee, S.I., and Batzoglou, S. (2003). Application of
independent component analysis to microarrays.
Genome Biol., 4, R76.
Martinez, D., and Bray, A. (2003) Nonlinear Blind Source
Separation Using Kernels. IEEE Tr. on Neural
Networks, 14, 228-235.
Mischak, H., et al. (2009). Capillary electrophoresis-mass
spectrometry as powerful tool in biomarker discovery
and clinical diagnosis: an update of recent
developments. Mass Spectrom. Rev., 28, 703-724.
Peng, H., Long, F., and Ding, C. (2005). Feature selection
based on mutual information: criteria for max-
dependency, max-relevance and min-redundancy.
IEEE Tr. Pat. Anal. Mach. Intel., 27, 1226-1238.
Petricoin, E.F., et al. (2002a) .Use of proteomic patterns in
serum to identify ovarian cancer. The Lancet, 359,
572-577.
Petricoin, E.F., et al. (2002b). Serum proteomic patterns
for detection of prostate cancer. J. Natl. Canc.
Institute, 94, 1576-1578.
Reju, V. G., Koh, S. N., Soon, I. Y. (2009). An algorithm
for mixing matrix estimation in instantaneous blind
source separation. Sig. Proc., 89, 1762-1773.
Schachtner, R., et al. (2008). Knowledge-based gene
expression classification via matrix factorization.
Bioinformatics, 24, 1688-1697.
Schölkopf, B., and Smola, A. (2002). Learning with
kernels, The MIT Press, Cambridge, MA.
Shipp, M. A., et al. (2002). Diffuse large B-cell lymphoma
outcome prediction by gene expression profiling and
supervised machine learning. Nature Med., 8, 68-74.
Singh, D., et al. (2002). Gene expression correlates of
clinical prostate cancer behavior. Cancer Cell, 1, 203-
209.
Sprites, P., Glymour, C., and Scheines, R. (2000).
Causation, prediction, and search. The MIT Press,
2nd edition.
Stadtlthanner, K., et al.
(2008). Hybridizing Sparse
Component Analysis with Genetic Algorithms for
Microarray Analysis. Neurocomputing, 71, 2356-
2376.
Statnikov, A., et al. (2005a). A comprehensive evaluation
of multicategory classification methods for microarray
gene expression cancer diagnosis. Bioinformatics, 21
631-643.
Statnikov, A., et al. (2005b). GEMS: A system for
automated cancer diagnosis and biomarker discovery
from microarray gene expression data. Int. J. Med.
Informatics, 74, 491-503.
Vapnik, V. (1998). Statistical learning theory. Wiley-
Interscience, New York.
Yuh, C. H., Bolouri, H., and Davidson, E. H (1998).
Genomic cis-regulatory logic: experimental and
computational analysis of a sea urchin gene. Science,
279, 1896-1902.
BIOINFORMATICS2015-InternationalConferenceonBioinformaticsModels,MethodsandAlgorithms
92
